Scientific Computing
Numerical simulation of real-world phenomena provides fertile ground for building interdisciplinary relationships. The SCI Institute has a long tradition of building these relationships in a win-win fashion – a win for the theoretical and algorithmic development of numerical modeling and simulation techniques and a win for the discipline-specific science of interest. High-order and adaptive methods, uncertainty quantification, complexity analysis, and parallelization are just some of the topics being investigated by SCI faculty. These areas of computing are being applied to a wide variety of engineering applications ranging from fluid mechanics and solid mechanics to bioelectricity.
Valerio Pascucci
Scientific Data Management
Chris Johnson
Problem Solving Environments
Ross Whitaker
GPUs
Chuck Hansen
GPUsFunded Research Projects:
Publications in Scientific Computing:
  Real-time magnetic resonance imaging-guided radiofrequency atrial ablation and visualization of lesion formation at 3 Tesla G.R. Vergara, S. Vijayakumar, E.G. Kholmovski, J.J. Blauer, M.A. Guttman, C. Gloschat, G. Payne, K. Vij, N.W. Akoum, M. Daccarett, C.J. McGann, R.S. Macleod, N.F. Marrouche. In Heart Rhythm, Vol. 8, No. 2, pp. 295--303. 2011. PubMed ID: 21034854 |
  Association of left atrial fibrosis detected by delayed-enhancement magnetic resonance imaging and the risk of stroke in patients with atrial fibrillation M. Daccarett, T.J. Badger, N. Akoum, N.S. Burgon, C. Mahnkopf, G.R. Vergara, E.G. Kholmovski, C.J. McGann, D.L. Parker, J. Brachmann, R.S. Macleod, N.F. Marrouche. In Journal of the American College of Cardiology, Vol. 57, No. 7, pp. 831--838. 2011. PubMed ID: 21310320 |
  MRI of the left atrium: predicting clinical outcomes in patients with atrial fibrillation M. Daccarett, C.J. McGann, N.W. Akoum, R.S. MacLeod, N.F. Marrouche. In Expert Review of Cardiovascular Therapy, Vol. 9, No. 1, pp. 105--111. 2011. PubMed ID: 21166532 |
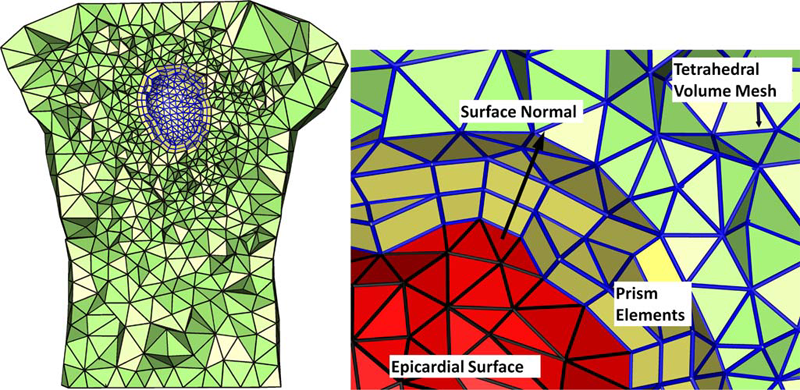
  Finite Element Based Discretization and Regularization Strategies for 3D Inverse Electrocardiography D. Wang, R.M. Kirby, C.R. Johnson. In IEEE Transactions for Biomedical Engineering, Vol. 58, No. 6, pp. 1827--1838. 2011. PubMed ID: 21382763 PubMed Central ID: PMC3109267 We consider the inverse electrocardiographic problem of computing epicardial potentials from a body-surface potential map. We study how to improve numerical approximation of the inverse problem when the finite-element method is used. Being ill-posed, the inverse problem requires different discretization strategies from its corresponding forward problem. We propose refinement guidelines that specifically address the ill-posedness of the problem. The resulting guidelines necessitate the use of hybrid finite elements composed of tetrahedra and prism elements. Also, in order to maintain consistent numerical quality when the inverse problem is discretized into different scales, we propose a new family of regularizers using the variational principle underlying finite-element methods. These variational-formed regularizers serve as an alternative to the traditional Tikhonov regularizers, but preserves the L2 norm and thereby achieves consistent regularization in multiscale simulations. The variational formulation also enables a simple construction of the discrete gradient operator over irregular meshes, which is difficult to define in traditional discretization schemes. We validated our hybrid element technique and the variational regularizers by simulations on a realistic 3-D torso/heart model with empirical heart data. Results show that discretization based on our proposed strategies mitigates the ill-conditioning and improves the inverse solution, and that the variational formulation may benefit a broader range of potential-based bioelectric problems. |
  A Diffusion Approach to Network Localization Y. Keller, Y. Gur. In IEEE Transactions on Signal Processing, Vol. 59, No. 6, pp. 2642--2654. 2011. DOI: 10.1109/TSP.2011.2122261 |
  From h to p Efficiently: Strategy Selection for Operator Evaluation on Hexahedral and Tetrahedral Elements C.D. Cantwell, S.J. Sherwin, R.M. Kirby, P.H.J. Kelly. In Computers and Fluids, Vol. 43, No. 1, pp. 23--28. 2011. DOI: 10.1016/j.compfluid.2010.08.012 |
  Finite element modeling of subcutaneous implantable defibrillator electrodes in an adult torso M. Jolley, J. Stinstra, J. Tate, S. Pieper, R.S. Macleod, L. Chu, P. Wang, J.K. Triedman. In Heart Rhythm, Vol. 7, No. 5, pp. 692--698. May, 2010. DOI: 10.1016/j.hrthm.2010.01.030 PubMed ID: 20230927 PubMed Central ID: PMC3103844 BACKGROUND: We used image-based finite element models (FEM) to predict the myocardial electric field generated during defibrillation shocks (pseudo-DFT) in a wide variety of reported and innovative subcutaneous electrode positions to determine factors affecting optimal lead positions for subcutaneous implantable cardioverter-defibrillators (S-ICD). METHODS: An image-based FEM of an adult man was used to predict pseudo-DFTs across a wide range of technically feasible S-ICD electrode placements. Generator location, lead location, length, geometry and orientation, and spatial relation of electrodes to ventricular mass were systematically varied. Best electrode configurations were determined, and spatial factors contributing to low pseudo-DFTs were identified using regression and general linear models. RESULTS: A total of 122 single-electrode/array configurations and 28 dual-electrode configurations were simulated. Pseudo-DFTs for single-electrode orientations ranged from 0.60 to 16.0 (mean 2.65 +/- 2.48) times that predicted for the base case, an anterior-posterior configuration recently tested clinically. A total of 32 of 150 tested configurations (21%) had pseudo-DFT ratios /=1, indicating the possibility of multiple novel, efficient, and clinically relevant orientations. Favorable alignment of lead-generator vector with ventricular myocardium and increased lead length were the most important factors correlated with pseudo-DFT, accounting for 70% of the predicted variation (R(2) = 0.70, each factor P < .05) in a combined general linear modl in which parameter estimates were calculated for each factor. CONCLUSION: Further exploration of novel and efficient electrode configurations may be of value in the development of the S-ICD technologies and implant procedure. FEM modeling suggests that the choice of configurations that maximize shock vector alignment with the center of myocardial mass and use of longer leads is more likely to result in lower DFT. |
  Quantifying Variability in Radiation Dose Due to Respiratory-Induced Tumor Motion S.E. Geneser, J.D. Hinkle, R.M. Kirby, Brian Wang, B. Salter, S. Joshi. In Medical Image Analysis, Vol. 15, No. 4, pp. 640--649. 2010. DOI: 10.1016/j.media.2010.07.003 |
  Towards the Development on an h-p-Refinement Strategy Based Upon Error Estimate Sensitivity P.K. Jimack, R.M. Kirby. In Computers and Fluids, Vol. 46, No. 1, pp. 277--281. 2010. DOI: 10.1016/j.compfluid.2010.08.003 The use of (a posteriori) error estimates is a fundamental tool in the application of adaptive numerical methods across a range of fluid flow problems. Such estimates are incomplete however, in that they do not necessarily indicate where to refine in order to achieve the most impact on the error, nor what type of refinement (for example h-refinement or p-refinement) will be best. This paper extends preliminary work of the authors (Comm Comp Phys, 2010;7:631–8), which uses adjoint-based sensitivity estimates in order to address these questions, to include application with p-refinement to arbitrary order and the use of practical a posteriori estimates. Results are presented which demonstrate that the proposed approach can guide both the h-refinement and the p-refinement processes, to yield improvements in the adaptive strategy compared to the use of more orthodox criteria. |
  Incorporating patient breathing variability into a stochastic model of dose deposition for stereotactic body radiation therapy S.E. Geneser, R.M. Kirby, Brian Wang, B. Salter, S. Joshi. In Information Processing in Medical Imaging, Lecture Notes in Computer Science LNCS, Vol. 5636, pp. 688--700. 2009. PubMed ID: 19694304 |