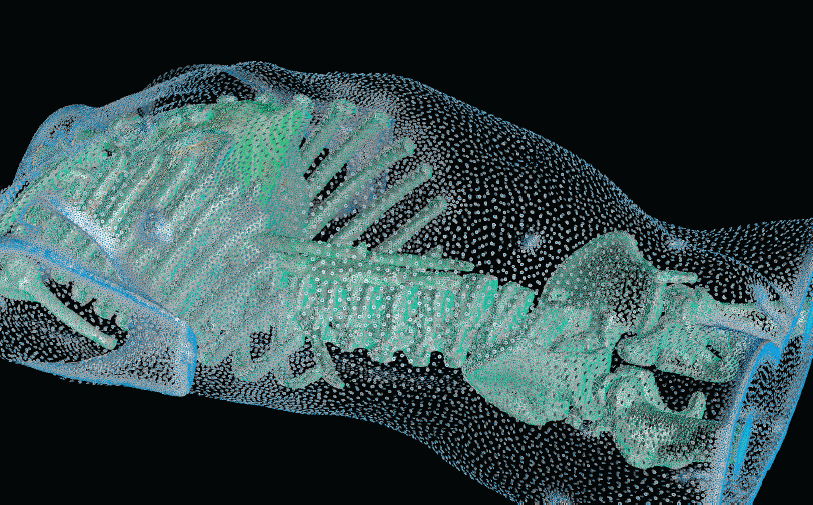
With the widespread use of medical imaging, there is a growing need for better analysis of datasets. One method for improving analysis is to simulate biological processes and medical interventions in silico, in order to render better predictions. For example, the CIBC center is currently collaborating with Dr. Triedman at Children's Hospital in Boston to develop a computer model that will help guide the implantation of Implantable Cardiac Defibrillators (ICDs). This model uses pediatric imaging to select placement of electrode leads to generate the optimal field for defibrillation. One of the critical pieces in the development of the model is the generation of quality meshes for electric field simulation. Because the project is entering the validation phase where many cases need to be reviewed, a robust and automated Meshing Pipeline is required.
Whether the object of analysis is the electrical stimulation of the brain, the defibrillation of the heart, the prediction of blood flow through the body, or the mechanical properties of bones, the need for quality meshes remains a common thread. In this research highlight, we describe some of our recent progress in creating and deploying our Meshing Pipeline system, BioMesh3D.
The goal of the BioMesh3D is to construct high-quality polygonal meshes from image-based segmentations; progress in the past year includes the first release of BioMesh3D as part of SCIRun 4.3. The approach in BioMesh3D is to assemble components from SCIRun and third party, open-source projects and to generate meshes in a range of formats. By basing BioMesh3D on SCIRun, there is an additional benefit of making use of many existing modules, thus allowing a rapid prototyping and testing process.
Like our other software projects, BioMesh3D development is driven by collaborations. Primary among them is the creation of patient-specific models of human torsos for use in simulating defibrillation. We also receive samples of explanted hearts in this context, and we are creating high resolution models that include fiber architecture from the diffusion weighted MRI scans.
A "typical" workflow that applies to many problems in biomedical simulation contains the following elements:
- Image acquisition and processing for a tissue, organ, or region of interest (imaging and image processing);
- Identification of structures, tissues, cells, or organelles within the images (image processing and segmentation);
- Fitting geometric surfaces to the boundaries between structures and regions (geometric modeling);
- Generation of three-dimensional volume mesh from hexahedra or tetrahedra (meshing); and
- Application of tissue parameters and boundary conditions and computation of spatial distribution of scalar, vector, or tensor quantities of interest (simulation).
BioMesh3D Software
 The BioMesh3D project aims to develop an easy-to-use program for generating quality meshes for use in biological simulations. The project's primary goal is to provide a solution that automatically generates a proper mesh and does not require much interaction with the user.
The BioMesh3D project aims to develop an easy-to-use program for generating quality meshes for use in biological simulations. The project's primary goal is to provide a solution that automatically generates a proper mesh and does not require much interaction with the user.To run the system, users only need a light-weight client application – all of the computationally expensive work is done on the servers. When users launch the client application, they login, which connects them to a special server called the Services Broker. All logins are secure (login and group information), and a list of datasets is stored in the SQLite database. The Services Broker shows the user what label-maps they've uploaded so far, and what meshes they've created (or have started creating). Users can upload a new label-map or start working with an existing one. For each mesh a user generates, there is a set of configuration parameters – reasonable defaults are created based on the uploaded label-map, and of course the user can override these. When the final tetrahedral mesh is generated, users can download it to their local machine. For many of the pipeline stages, there's an interactive visualization that the user can run to explore and analyze results.
Visualizations are rendered on the server – as the user navigates around, an MPEG stream is sent to the client app for display. In addition to providing light-weight client app binaries (and source), CIBC makes the server available to people who want to install and run it on their local servers.
Current Research Progress
BioMesh3D is built on a pipeline on image acquisition and segmentation, interface surface detection, sizing field computation, particle distribution, and finally, mesh generation. One of the more complex stages in the pipeline involves estimating an appropriate sizing field for the non-smooth, non-manifold geometries of the interface surfaces that bind regions of different materials. The sizing field represents a radius of influence for each particle within the mesh. In BioMesh3D, a computation of the medial axis is required, as well as processing to tighten the medial axis and regularize the sizing field. An appropriate sizing field must capture both the topological and geometric features of the interface surface in an adaptive way. In general, to satisfy these needs, expensive, global computations are required.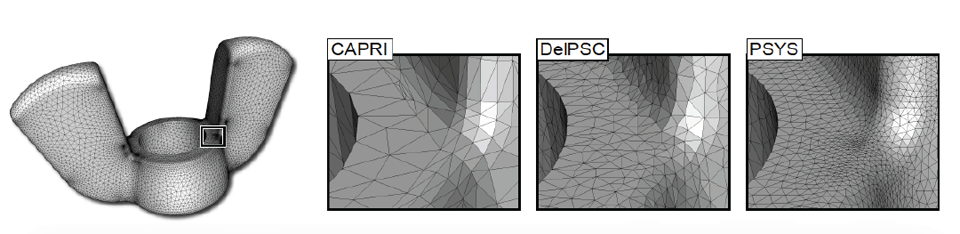 |
| Figure from J. Bronson, et al., Particle Systems for Adaptive, Isotropic Meshing of CAD Models. WingNut mesh for PSYS. WingNut mesh for PSYS. Insets show comparison between CAPRI, DelPSC, and PSYS. |
Recently, we have worked on a particle system technique where sizing field information is estimated using only local information. This project, designed for CAD inputs common in engineering fields, computes a sizing field within the particle system optimization by allowing particles to estimate the radius based on their current neighborhood information. In particular, each particle tries to adapt to geometric features (curvature), enforce topological constraints (separation from particles on distinct topological patches), and have a sizing field which does not vary too far from that of its neighbors (adaptivity and continuity conditions). While the particle system solves a more complex energy minimization, the exchange eliminates the need for a global sizing field computation.
The results of our work have been well received by the meshing community because of the synthesis of cutting-edge meshing techniques. The algorithm and software package, PSYS, received the Best Paper award at the 19th International Meshing Roundtable in Chattanooga, TN. We are optimistic that a similar approach can be translated back from CAD domain inputs to biological, image-based inputs, leading to computational savings in one of the more expensive stages of the BioMesh3D pipeline.
MeshMed Workshop
This coming year, CIBC members are organizing a workshop on Mesh Processing in Medical Image Analysis, held in conjuncture with MICCAI 2011 in Toronto. This is the first workshop on meshing in Medical Imaging, which has the goal to gather those who create the data, those who program the mesh, and those who employ the meshes. This workshop proposes to improve the cross-pollination of the imaging and meshing efforts by considering how meshing fits into the end-to-end pipeline from image acquisition to clinical analysis.References
J.R. Bronson, J.A. Levine, R.T. Whitaker. "Particle Systems for Adaptive, Isotropic Meshing of CAD Models," In Proceedings of the 19th International Meshing Roundtable, Chattanooga, TN, Note: Awarded Best Paper, pp. (to appear). October, 2010.M. Callahan, M.J. Cole, J.F. Shepherd, J.G. Stinstra, C.R. Johnson. "A Meshing Pipeline for Biomedical Models," In Engineering with Computers, Vol. 25, No. 1, Springer Link, pp. 115-130. 2009.
M. Meyer, R.T. Whitaker, R.M. Kirby, C. Ledergerber, H. Pfister. "Particle-based Sampling and Meshing of Surfaces in Multimaterial Volumes," In IEEE Transactions on Visualization and Computer Graphics, Vol. 14, No. 6, pp. 1539--1546. 2008.
M. Meyer, R.M. Kirby, R.T. Whitaker. "Topology, Accuracy, and Quality of Isosurface Meshes Using Dynamic Particles," In Proceedings of Visualization 2007 (IEEE TVCG), Vol. 13, No. 6, pp. 1704--1711. 2007