"I am excited about the SCIRun and BioPSE software releases," says the Institute's director Christopher R. Johnson. "We have been working on SCIRun since 1992. What started out as software designed by a few people (Steve Parker with help from David Weinstein) has grown into a substantial software effort with more than 50 contributors." Johnson continues, "BioPSE has been a focused software effort since 1999. I am thrilled to see SCIRun and BioPSE released and look forward to seeing how scientists and engineers use these software packages to solve their application domain problems."
The release of SCIRun/BioPSE is marked with a great deal of enthusiasm from its collaborators. The SCI Institute anticipates exponential growth in the functionality of the software, and its application to the medical community. SCIRun/BioPSE is a leap toward bridging the gap between research and surgical procedures, allowing the physician to interactively view three-dimensional data from MRI, MRA, CT, MEG, EEG, and PET scans.
What are SCIRun and BioPSE?
SCIRun
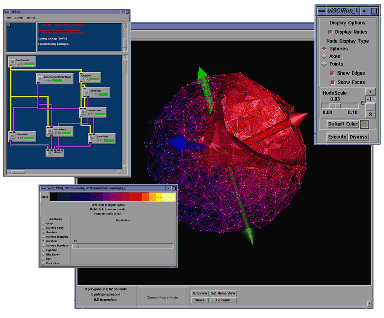
Simply put, SCIRun is a multifunctional problem solving environment that can be best described as a computational workbench by which the user can "close the loop." All aspects of the modeling, simulation, and visualization processes are linked, controlled graphically within the context of a single application program. Previous approaches to solving computational science problems involved a predominantly batch-mode, off-line, non-interactive computational science pipeline where the organizing, running and visualizing of new large-scale simulations required hours or even days of a researcher's time. In order to obtain immediate results, more efficient methods were needed.Creating an integrated problem solving environment for scientific computing involves many significant elements: representing mathematical and geometrical models; computational solution of the governing equations and visualization of models and results--all within an efficient, parallel software environment. By tackling such a large integrated systems approach, the SCI Institute draws upon its multidisciplinary approach to science and research in geometric modeling, simulation, scientific visualization and software environments.
BioPSE
In 1998, the SCI Institute submitted a grant proposal to the NIH/NCRR to develop deliverable and clinically usable software under the name BioPSE (as well as other software projects). Under the grant, BioPSE would be a collection of algorithms used to run bioelectric field simulations inside of SCIRun. To that point, SCIRun had been used as a prototyping platform for research ideas by students and staff in the center and, as such, was largely comprised of proof of concept style code rather than formally engineered, deliverable style code. These elements, combined with the fact that most of the SCI Institute staff were from academia rather than industry, provided a great challenge for bringing SCIRun and BioPSE to the general public.How are SCIRun and BioPSE related?
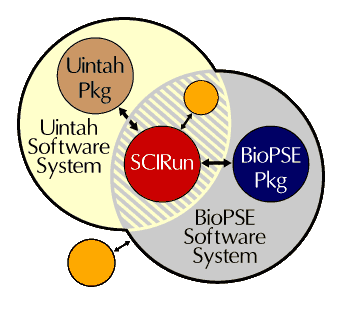
In addition to the major projects that have both leveraged and advanced SCIRun, a number of smaller packages exist, extending SCIRun's utility. Examples include the Teem package for raster data processing; the NetSolve package for linear algebra subroutines (developed by researchers at the University of Tennessee and Knoxville); and a communications interface, bridging SCIRun and Matlab environments. The SCI Institute has developed various forms of software wrappers or interfaces that allow SCIRun to leverage the strengths of these third party tools. These links are referred to as "bridges."
There are also instances in which a tighter level of integration rather than a bridge between SCIRun and third-party software is necessary. One such example is the addition of mpeg support for capturing animations from the SCIRun Viewer module that uses the Berkeley and Alex Knowles' mpeg encoding tools. Another example is Paul Haeberli's Libimage, a set of image generation and manipulation tools. To indicate whether or not such tools are available, the configure scripts for SCIRun contain optional control flags.
The SCI Institute believes that this combination of a robust infrastructure and modular extensibility through packages and third-party libraries will allow SCIRun to grow and adapt to scientist's changing needs and opportunities.
