Bayesian Source Imaging of Pediatric Epilepsy
Collaborating Investigator: Simon Warfield, Director of the Computational Radiology LaboratoryEpilepsy is a brain disorder in which a person has repeated seizures (convulsions) over time. It affects over 2.5 million Americans and has an estimated total annual health care cost close to $12.5 billion per year. Medications for epilepsy often have significant side effects and currently fail to halt seizures in up to 20% of patients. Approximately 75% of epilepsy patients have their first seizure in childhood. Many only become candidates for surgical intervention after a long period of partially-effective medication that can have debilitating educational and sociological side effects. The overall objective of this DBP is to develop quantitative analysis algorithms to dramatically improve our capacity to detect and localize epileptogenic foci in brain tissue, in order to enable curative surgery for more patients. Technological innovations offer the prospect of improving foci localization and thereby patient-specific treatment. The treatment may be successful by resection or transsecting white matter fiber pathways in the vicinity of epileptic foci. These assessments rely on knowing where the seizure foci are located and their relationship to surrounding structures to prevent damage of brain functionality. With poor localization, some patients may be considered inoperable. If more accurate foci localization can be developed, more patients will be considered candidates for surgery. Today, the use of sophisticated physics and realistic forward models has been made practical by the exponentially increasing computing capacity of modern workstations. Furthermore, the use of computationally expensive methods can dramatically improve the reliability to localize epileptic foci (inverse solution), enabling more realistic models and allowing better noise and artifact rejection. Unfortunately, these technologies have not yet reached clinical practice. Each patient who is identified by the clinical epilepsy service for evaluation as a potential candidate for surgery will be approached and given the opportunity to participate in this study. If a patient chooses to participate in the study, additional data acquisition and data analysis will be carried out. All patients will receive the standard clinical care and independent assessment. Experts blinded to the clinical interpretation of the data will review fused multimodal data and determine if the patient is a candidate for surgical intervention based on the presence of a focal seizure zone.
The goal of this DBP is to develop algorithms to improve the detection of epilepsy using electroencephalography (EEG) and to localize its origin in the human brain. To achieve this goal, epileptic spikes need to be identified based on measured EEG data and disentangled from other signal sources. After isolating signals related to epileptic brain activity, source localization (inverse problem), which is based on advanced volume conductor modeling (forward problem), can be employed. As a prerequisite to research on this topic we and our collaborators applied for and received permission to work on epileptic patient data from the institutional review boards (IRB) of Boston Children's Hospital and the University of Utah.
Isolating Epileptic Brain Activity from Surface EEG using Laplacian Eigenmaps
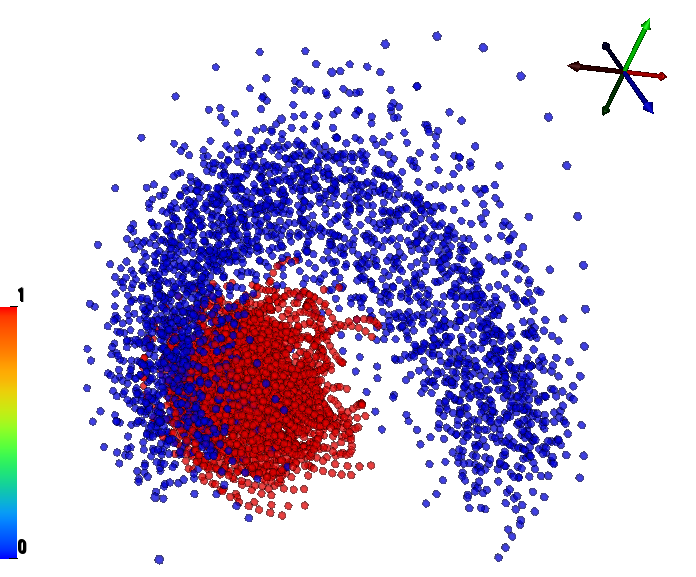
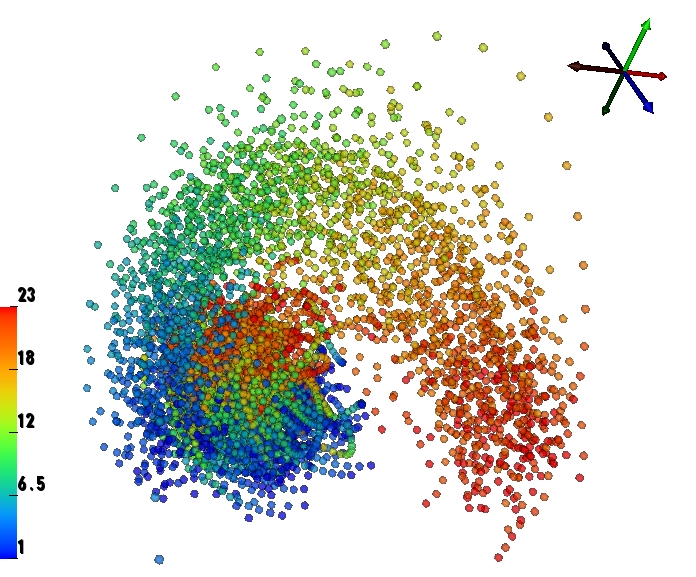
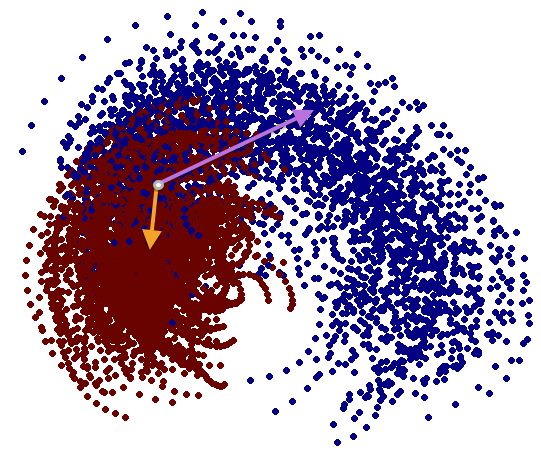
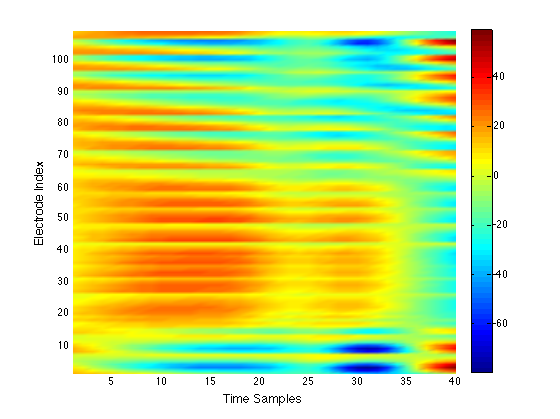
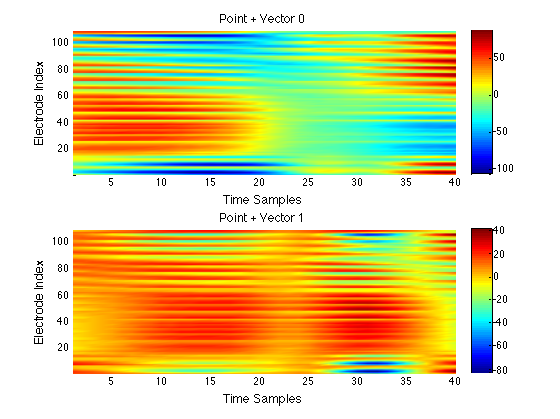
We have performed a preliminary investigation of the relationship between the dynamics in EEG data recorded from a subject during normal background activity and epileptic brain activity. We identified two classes of spikes from the frequent episodes of interictal spiking activity. Therefore, we applied a number of novel nonlinear mapping methods based on Laplacian eigenmaps (LE). LE is a well-known nonlinear dimensionality technique from the machine learning literature and is used here to analyze the differences between background activity data and spiking activity data. The analysis may elucidate relationships between data from different electrodes (not just between background and spiking activity) and also between the two classes of spiking activity. In the figure below, we show two visualizations of the same data segments containing spiking that have been mapped to a three-dimensional subspace of the LE output space. We use color to show the spike class (top) and time relative to the start of each data segment (bottom). In In the figure, we picked a representative point from the LE space where the two spike classes overlap. The figure further shows that the point corresponds to a spatial and temporal map containing both "slow waves" (long ellipsoidal blue regions) and "theta bursts" (shorter ellipsoidal blue regions). Using the differential of the LE function we mapped vectors pointing at the centers of the two spike class clusters back to the electrode space. The added vector also isolates the corresponding spike type in the map.
Localization of Epileptic Foci from EEG Measurements
In order to perform source localization a description of electrical properties (a volume conductor model) of the patient's head needs to be generated. We can then compute the contribution to EEG measurements of a given electrode configuration and potential source locations in the brain. (forward problem). We generated volume conductor models for the patient and evaluated several modeling aspects. First, we improved the segmentation of tissue boundaries, which were generated from automatic segmentation procedures. Second, we used two meshing packages (BioMesh3D1, Cleaver2) to evaluate the quality of the resulting tetrahedral mesh representing spatial features of the modeled tissues. The results indicate that Cleaver performed well compared to BioMesh3D in many aspects, such as speed to generate a mesh and mesh quality considerations, which have an impact on the stability of numerical simulations. Third, we computed finite element solutions (FEM) and compared them to finite difference solutions (FDM) provided by S. Warfield's group. Since conformal tissue boundaries are preserved using Cleaver and due to the fact that we want to model anisotropic inhomogeneous conductivity distributions, we decided to use FEM over FDM. In a next step we are going to localize epileptic activity-related signals that are provided by the LE method. We plan to use common discrete and distributed source localization methods. The comparison of source localization results to more sophisticated methods (Bayesian source localization and specifically adapted methods for LE projections) together with clinical expertise might give us more insight into different classes of epileptic spikes, foci location, and their development over time.References:
1. M.D. Meyer, R.T. Whitaker, R.M. Kirby, C. Ledergerber, and H. Pfister. Particle-based sampling and meshing of surfaces in multimaterial volumes. IEEE Transactions on Visualization and Computer Graphics, 14(6):1539–1546, 2008.2. J.R. Bronson, J.A. Levine, and R.T. Whitaker. Lattice cleaving: Conforming tetrahedral meshes of multimaterial domains with bounded quality. In Proceedings of the 21st International Meshing Roundtable, pages 191–209, 2012.