SCI Publications
2023


S. Leventhal, A. Gyulassy, M. Heimann, V. Pascucci.
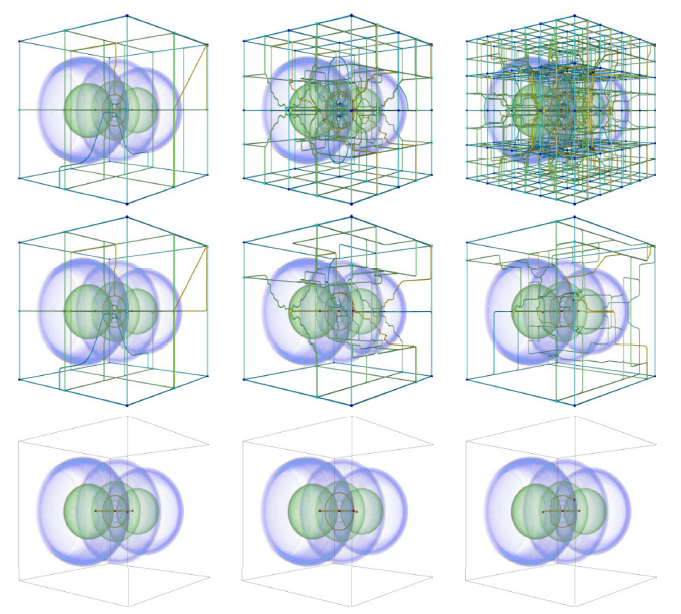
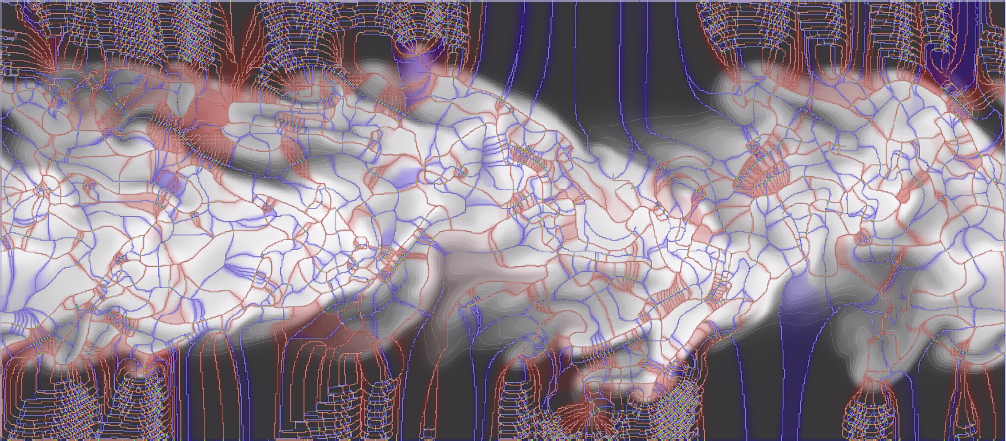
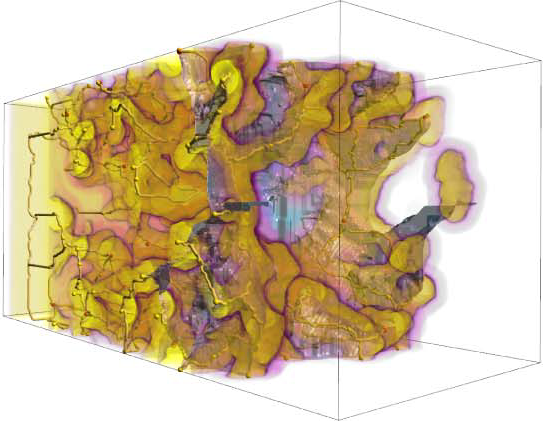
“Exploring Classification of Topological Priors with Machine Learning for Feature Extraction,” In IEEE Transactions on Visualization and Computer Graphics, pp. 1--12. 2023.
In many scientific endeavors, increasingly abstract representations of data allow for new interpretive methodologies and conceptualization of phenomena. For example, moving from raw imaged pixels to segmented and reconstructed objects allows researchers new insights and means to direct their studies toward relevant areas. Thus, the development of new and improved methods for segmentation remains an active area of research. With advances in machine learning and neural networks, scientists have been focused on employing deep neural networks such as U-Net to obtain pixel-level segmentations, namely, defining associations between pixels and corresponding/referent objects and gathering those objects afterward. Topological analysis, such as the use of the Morse-Smale complex to encode regions of uniform gradient flow behavior, offers an alternative approach: first, create geometric priors, and then apply machine learning to classify. This approach is empirically motivated since phenomena of interest often appear as subsets of topological priors in many applications. Using topological elements not only reduces the learning space but also introduces the ability to use learnable geometries and connectivity to aid the classification of the segmentation target. In this paper, we describe an approach to creating learnable topological elements, explore the application of ML techniques to classification tasks in a number of areas, and demonstrate this approach as a viable alternative to pixel-level classification, with similar accuracy, improved execution time, and requiring marginal training data.
2022


Y. Livnat, D. Maljovec, A. Gyulassy, B. Mouginot, V. Pascucci.
“A Novel Tree Visualization to Guide Interactive Exploration of Multi-dimensional Topological Hierarchies,” Subtitled “arXiv preprint arXiv:2208.06952,” 2022.
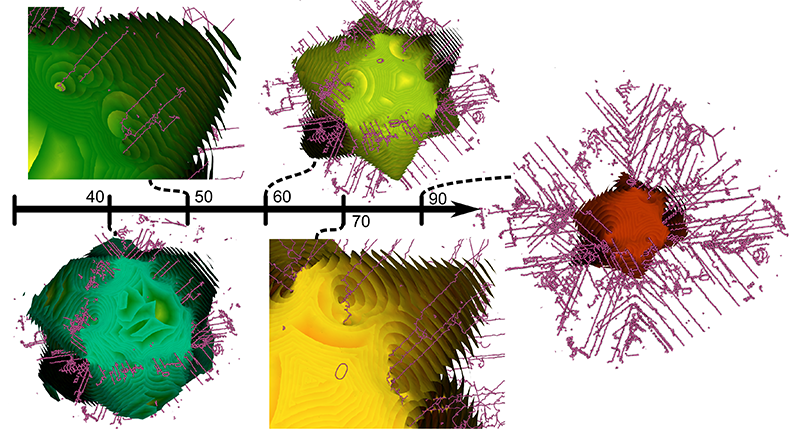
Understanding the response of an output variable to multi-dimensional inputs lies at the heart of many data exploration endeavours. Topology-based methods, in particular Morse theory and persistent homology, provide a useful framework for studying this relationship, as phenomena of interest often appear naturally as fundamental features. The Morse-Smale complex captures a wide range of features by partitioning the domain of a scalar function into piecewise monotonic regions, while persistent homology provides a means to study these features at different scales of simplification. Previous works demonstrated how to compute such a representation and its usefulness to gain insight into multi-dimensional data. However, exploration of the multi-scale nature of the data was limited to selecting a single simplification threshold from a plot of region count. In this paper, we present a novel tree visualization that provides a concise overview of the entire hierarchy of topological features. The structure of the tree provides initial insights in terms of the distribution, size, and stability of all partitions. We use regression analysis to fit linear models in each partition, and develop local and relative measures to further assess uniqueness and the importance of each partition, especially with respect parents/children in the feature hierarchy. The expressiveness of the tree visualization becomes apparent when we encode such measures using colors, and the layout allows an unprecedented level of control over feature selection during exploration. For instance, selecting features from multiple scales of the hierarchy enables a more nuanced exploration. Finally, we …


A. Venkat, D. Hoang, A. Gyulassy, P.T. Bremer, F. Federer, V. Pascucci.
“High-Quality Progressive Alignment of Large 3D Microscopy Data,” In 2022 IEEE 12th Symposium on Large Data Analysis and Visualization (LDAV), pp. 1--10. 2022.
DOI: 10.1109/LDAV57265.2022.9966406
Large-scale three-dimensional (3D) microscopy acquisitions fre-quently create terabytes of image data at high resolution and magni-fication. Imaging large specimens at high magnifications requires acquiring 3D overlapping image stacks as tiles arranged on a two-dimensional (2D) grid that must subsequently be aligned and fused into a single 3D volume. Due to their sheer size, aligning many overlapping gigabyte-sized 3D tiles in parallel and at full resolution is memory intensive and often I/O bound. Current techniques trade accuracy for scalability, perform alignment on subsampled images, and require additional postprocess algorithms to refine the alignment quality, usually with high computational requirements. One common solution to the memory problem is to subdivide the overlap region into smaller chunks (sub-blocks) and align the sub-block pairs in parallel, choosing the pair with the most reliable alignment to determine the global transformation. Yet aligning all sub-block pairs at full resolution remains computationally expensive. The key to quickly developing a fast, high-quality, low-memory solution is to identify a single or a small set of sub-blocks that give good alignment at full resolution without touching all the overlapping data. In this paper, we present a new iterative approach that leverages coarse resolution alignments to progressively refine and align only the promising candidates at finer resolutions, thereby aligning only a small user-defined number of sub-blocks at full resolution to determine the lowest error transformation between pairwise overlapping tiles. Our progressive approach is 2.6x faster than the state of the art, requires less than 450MB of peak RAM (per parallel thread), and offers a higher quality alignment without the need for additional postprocessing refinement steps to correct for alignment errors.
2021


A. A. Gooch, S. Petruzza, A. Gyulassy, G. Scorzelli, V. Pascucci, L. Rantham, W. Adcock, C. Coopmans.
“Lessons learned towards the immediate delivery of massive aerial imagery to farmers and crop consultants,” In Autonomous Air and Ground Sensing Systems for Agricultural Optimization and Phenotyping VI, Vol. 11747, International Society for Optics and Photonics, pp. 22 -- 34. 2021.
DOI: 10.1117/12.2587694
In this paper, we document lessons learned from using ViSOAR Ag Explorer™ in the fields of Arkansas and Utah in the 2018-2020 growing seasons. Our insights come from creating software with fast reading and writing of 2D aerial image mosaics for platform-agnostic collaborative analytics and visualization. We currently enable stitching in the field on a laptop without the need for an internet connection. The full resolution result is then available for instant streaming visualization and analytics via Python scripting. While our software, ViSOAR Ag Explorer™ removes the time and labor software bottleneck in processing large aerial surveys, enabling a cost-effective process to deliver actionable information to farmers, we learned valuable lessons with regard to the acquisition, storage, viewing, analysis, and planning stages of aerial data surveys. Additionally, with the ultimate goal of stitching thousands of images in minutes on board a UAV at the time of data capture, we performed preliminary tests for on-board, real-time stitching and analysis on USU AggieAir sUAS using lightweight computational resources. This system is able to create a 2D map while flying and allow interactive exploration of the full resolution data as soon as the platform has landed or has access to a network. This capability further speeds up the assessment process on the field and opens opportunities for new real-time photogrammetry applications. Flying and imaging over 1500-2000 acres per week provides up-to-date maps that give crop consultants a much broader scope of the field in general as well as providing a better view into planting and field preparation than could be observed from field level. Ultimately, our software and hardware could provide a much better understanding of weed presence and intensity or lack thereof.


X. Huang, P. Klacansky, S. Petruzza, A. Gyulassy, P.T. Bremer, V. Pascucci.
“Distributed merge forest: a new fast and scalable approach for topological analysis at scale,” In Proceedings of the ACM International Conference on Supercomputing, pp. 367-377. 2021.
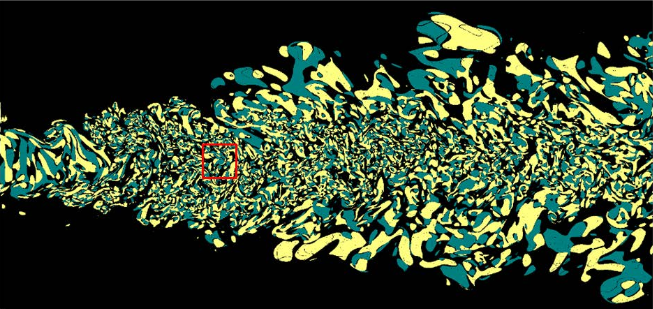
Topological analysis is used in several domains to identify and characterize important features in scientific data, and is now one of the established classes of techniques of proven practical use in scientific computing. The growth in parallelism and problem size tackled by modern simulations poses a particular challenge for these approaches. Fundamentally, the global encoding of topological features necessitates inter process communication that limits their scaling. In this paper, we extend a new topological paradigm to the case of distributed computing, where the construction of a global merge tree is replaced by a distributed data structure, the merge forest, trading slower individual queries on the structure for faster end-to-end performance and scaling. Empirically, the queries that are most negatively affected also tend to have limited practical use. Our experimental results demonstrate the scalability of both the merge forest construction and the parallel queries needed in scientific workflows, and contrast this scalability with the two established alternatives that construct variations of a global tree.


A. Venkat, A. Gyulassy, G. Kosiba, A. Maiti, H. Reinstein, R. Gee, P.-T. Bremer, V. Pascucci.
“Towards replacing physical testing of granular materials with a Topology-based Model,” Subtitled “arXiv preprint arXiv:2109.08777,” 2021.
In the study of packed granular materials, the performance of a sample (e.g., the detonation of a high-energy explosive) often correlates to measurements of a fluid flowing through it. The "effective surface area," the surface area accessible to the airflow, is typically measured using a permeametry apparatus that relates the flow conductance to the permeable surface area via the Carman-Kozeny equation. This equation allows calculating the flow rate of a fluid flowing through the granules packed in the sample for a given pressure drop. However, Carman-Kozeny makes inherent assumptions about tunnel shapes and flow paths that may not accurately hold in situations where the particles possess a wide distribution in shapes, sizes, and aspect ratios, as is true with many powdered systems of technological and commercial interest. To address this challenge, we replicate these measurements virtually on micro-CT images of the powdered material, introducing a new Pore Network Model based on the skeleton of the Morse-Smale complex. Pores are identified as basins of the complex, their incidence encodes adjacency, and the conductivity of the capillary between them is computed from the cross-section at their interface. We build and solve a resistive network to compute an approximate laminar fluid flow through the pore structure. We provide two means of estimating flow-permeable surface area: (i) by direct computation of conductivity, and (ii) by identifying dead-ends in the flow coupled with isosurface extraction and the application of the Carman-Kozeny equation, with the aim of establishing consistency over a range of particle shapes, sizes, porosity levels, and void distribution patterns.
2019


A. Gyulassy, P.-T. Bremer, V. Pascucci.
“Shared-Memory Parallel Computation of Morse-Smale Complexes with Improved Accuracy,” In IEEE Transactions on Visualization and Computer Graphics, Vol. 25, No. 1, IEEE, pp. 1183--1192. Jan, 2019.
DOI: 10.1109/tvcg.2018.2864848
Topological techniques have proven to be a powerful tool in the analysis and visualization of large-scale scientific data. In particular, the Morse-Smale complex and its various components provide a rich framework for robust feature definition and computation. Consequently, there now exist a number of approaches to compute Morse-Smale complexes for large-scale data in parallel. However, existing techniques are based on discrete concepts which produce the correct topological structure but are known to introduce grid artifacts in the resulting geometry. Here, we present a new approach that combines parallel streamline computation with combinatorial methods to construct a high-quality discrete Morse-Smale complex. In addition to being invariant to the orientation of the underlying grid, this algorithm allows users to selectively build a subset of features using high-quality geometry. In particular, a user may specifically select which ascending/descending manifolds are reconstructed with improved accuracy, focusing computational effort where it matters for subsequent analysis. This approach computes Morse-Smale complexes for larger data than previously feasible with significant speedups. We demonstrate and validate our approach using several examples from a variety of different scientific domains, and evaluate the performance of our method.
2018


S. Petruzza, A. Gyulassy, V. Pascucci,, P. T. Bremer.
“A Task-Based Abstraction Layer for User Productivity and Performance Portability in Post-Moore’s Era Supercomputing,” In 3RD INTERNATIONAL WORKSHOP ON POST-MOORE’S ERA SUPERCOMPUTING (PMES), 2018.
The proliferation of heterogeneous computing architectures in current and future supercomputing systems dramatically increases the complexity of software development and exacerbates the divergence of software stacks. Currently, task-based runtimes attempt to alleviate these impediments, however their effective use requires expertise and deep integration that does not facilitate reuse and portability. We propose to introduce a task-based abstraction layer that separates the definition of the algorithm from the runtime-specific implementation, while maintaining performance portability.


S. Petruzza, A. Gyulassy, V. Pascucci,, P. T. Bremer.
“A Task-Based Abstraction Layer for User Productivity and Performance Portability in Post-Moore’s Era Supercomputing,” In 3RD INTERNATIONAL WORKSHOP ON POST-MOORE’S ERA SUPERCOMPUTING (PMES), 2018.
The proliferation of heterogeneous computing architectures in current and future supercomputing systems dramatically increases the complexity of software development and exacerbates the divergence of software stacks. Currently, task-based runtimes attempt to alleviate these impediments, however their effective use requires expertise and deep integration that does not facilitate reuse and portability. We propose to introduce a task-based abstraction layer that separates the definition of the algorithm from the runtime-specific implementation, while maintaining performance portability.
2017


S. Petruzza, A. Venkat, A. Gyulassy, G. Scorzelli, F. Federer, A. Angelucci, V. Pascucci, P. T. Bremer.
“ISAVS: Interactive Scalable Analysis and Visualization System,” In ACM SIGGRAPH Asia 2017 Symposium on Visualization, ACM Press, 2017.
DOI: 10.1145/3139295.3139299
Modern science is inundated with ever increasing data sizes as computational capabilities and image acquisition techniques continue to improve. For example, simulations are tackling ever larger domains with higher fidelity, and high-throughput microscopy techniques generate larger data that are fundamental to gather biologically and medically relevant insights. As the image sizes exceed memory, and even sometimes local disk space, each step in a scientific workflow is impacted. Current software solutions enable data exploration with limited interactivity for visualization and analytic tasks. Furthermore analysis on HPC systems often require complex hand-written parallel implementations of algorithms that suffer from poor portability and maintainability. We present a software infrastructure that simplifies end-to-end visualization and analysis of massive data. First, a hierarchical streaming data access layer enables interactive exploration of remote data, with fast data fetching to test analytics on subsets of the data. Second, a library simplifies the process of developing new analytics algorithms, allowing users to rapidly prototype new approaches and deploy them in an HPC setting. Third, a scalable runtime system automates mapping analysis algorithms to whatever computational hardware is available, reducing the complexity of developing scaling algorithms. We demonstrate the usability and performance of our system using a use case from neuroscience: filtering, registration, and visualization of tera-scale microscopy data. We evaluate the performance of our system using a leadership-class supercomputer, Shaheen II.
2015

A. Gyulassy, A. Knoll, K. C. Lau, Bei Wang, P. T. Bremer, M. E. Papka, L. A. Curtiss, V. Pascucci.
“Morse-Smale Analysis of Ion Diffusion for DFT Battery Materials Simulations,” In Topology-Based Methods in Visualization (TopoInVis), 2015.
Ab initio molecular dynamics (AIMD) simulations are increasingly useful in modeling, optimizing and synthesizing materials in energy sciences. In solving Schrodinger's equation, they generate the electronic structure of the simulated atoms as a scalar field. However, methods for analyzing these volume data are not yet common in molecular visualization. The Morse-Smale complex is a proven, versatile tool for topological analysis of scalar fields. In this paper, we apply the discrete Morse-Smale complex to analysis of first-principles battery materials simulations. We consider a carbon nanosphere structure used in battery materials research, and employ Morse-Smale decomposition to determine the possible lithium ion diffusion paths within that structure. Our approach is novel in that it uses the wavefunction itself as opposed distance fields, and that we analyze the 1-skeleton of the Morse-Smale complex to reconstruct our diffusion paths. Furthermore, it is the first application where specific motifs in the graph structure of the complete 1-skeleton define features, namely carbon rings with specific valence. We compare our analysis of DFT data with that of a distance field approximation, and discuss implications on larger classical molecular dynamics simulations.


A. Gyulassy, A. Knoll, K. C. Lau, Bei Wang, PT. Bremer, M.l E. Papka, L. A. Curtiss, V. Pascucci.
“Interstitial and Interlayer Ion Diffusion Geometry Extraction in Graphitic Nanosphere Battery Materials,” In Proceedings IEEE Visualization Conference, 2015.
Large-scale molecular dynamics (MD) simulations are commonly used for simulating the synthesis and ion diffusion of battery materials. A good battery anode material is determined by its capacity to store ion or other diffusers. However, modeling of ion diffusion dynamics and transport properties at large length and long time scales would be impossible with current MD codes. To analyze the fundamental properties of these materials, therefore, we turn to geometric and topological analysis of their structure. In this paper, we apply a novel technique inspired by discrete Morse theory to the Delaunay triangulation of the simulated geometry of a thermally annealed carbon nanosphere. We utilize our computed structures to drive further geometric analysis to extract the interstitial diffusion structure as a single mesh. Our results provide a new approach to analyze the geometry of the simulated carbon nanosphere, and new insights into the role of carbon defect size and distribution in determining the charge capacity and charge dynamics of these carbon based battery materials.
2014


H. Bhatia, A. Gyulassy, H. Wang, P.-T. Bremer, V. Pascucci .
“Robust Detection of Singularities in Vector Fields,” In Topological Methods in Data Analysis and Visualization III, Mathematics and Visualization, Springer International Publishing, pp. 3--18. March, 2014.
DOI: 10.1007/978-3-319-04099-8_1
Recent advances in computational science enable the creation of massive datasets of ever increasing resolution and complexity. Dealing effectively with such data requires new analysis techniques that are provably robust and that generate reproducible results on any machine. In this context, combinatorial methods become particularly attractive, as they are not sensitive to numerical instabilities or the details of a particular implementation. We introduce a robust method for detecting singularities in vector fields. We establish, in combinatorial terms, necessary and sufficient conditions for the existence of a critical point in a cell of a simplicial mesh for a large class of interpolation functions. These conditions are entirely local and lead to a provably consistent and practical algorithm to identify cells containing singularities.


A.G. Landge, V. Pascucci, A. Gyulassy, J.C. Bennett, H. Kolla, J. Chen, P.-T. Bremer.
“In-situ feature extraction of large scale combustion simulations using segmented merge trees,” In Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis (SC 2014), New Orleans, Louisana, IEEE Press, Piscataway, NJ, USA pp. 1020--1031. 2014.
ISBN: 978-1-4799-5500-8
DOI: 10.1109/SC.2014.88
The ever increasing amount of data generated by scientific simulations coupled with system I/O constraints are fueling a need for in-situ analysis techniques. Of particular interest are approaches that produce reduced data representations while maintaining the ability to redefine, extract, and study features in a post-process to obtain scientific insights.
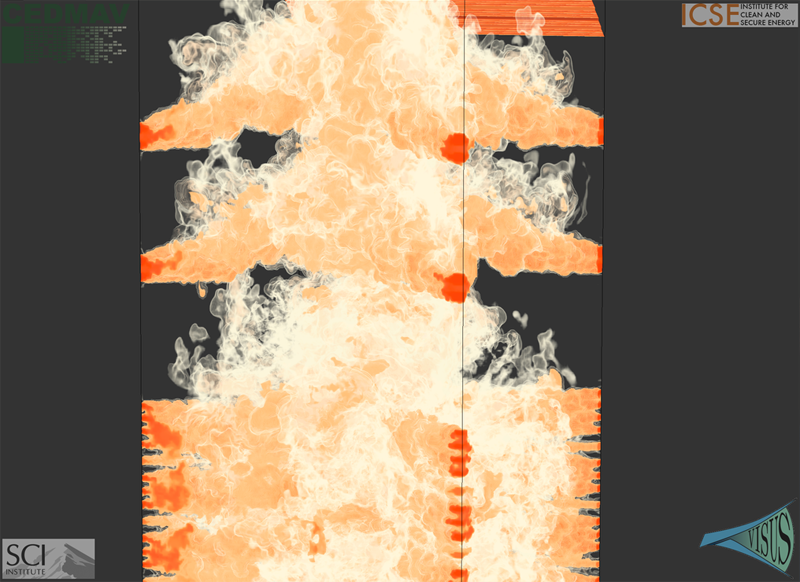
This paper presents two variants of in-situ feature extraction techniques using segmented merge trees, which encode a wide range of threshold based features. The first approach is a fast, low communication cost technique that generates an exact solution but has limited scalability. The second is a scalable, local approximation that nevertheless is guaranteed to correctly extract all features up to a predefined size. We demonstrate both variants using some of the largest combustion simulations available on leadership class supercomputers. Our approach allows state-of-the-art, feature-based analysis to be performed in-situ at significantly higher frequency than currently possible and with negligible impact on the overall simulation runtime.
2013


M. Gamell, I. Rodero, M. Parashar, J.C. Bennett, H. Kolla, J.H. Chen, P.-T. Bremer, A. Landge, A. Gyulassy, P. McCormick, Scott Pakin, Valerio Pascucci, Scott Klasky.
“Exploring Power Behaviors and Trade-offs of In-situ Data Analytics,” In Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis, Association for Computing Machinery, 2013.
ISBN: 978-1-4503-2378-9
DOI: 10.1145/2503210.2503303
As scientific applications target exascale, challenges related to data and energy are becoming dominating concerns. For example, coupled simulation workflows are increasingly adopting in-situ data processing and analysis techniques to address costs and overheads due to data movement and I/O. However it is also critical to understand these overheads and associated trade-offs from an energy perspective. The goal of this paper is exploring data-related energy/performance trade-offs for end-to-end simulation workflows running at scale on current high-end computing systems. Specifically, this paper presents: (1) an analysis of the data-related behaviors of a combustion simulation workflow with an in-situ data analytics pipeline, running on the Titan system at ORNL; (2) a power model based on system power and data exchange patterns, which is empirically validated; and (3) the use of the model to characterize the energy behavior of the workflow and to explore energy/performance trade-offs on current as well as emerging systems.
Keywords: SDAV


V. Pascucci, P.-T. Bremer, A. Gyulassy, G. Scorzelli, C. Christensen, B. Summa, S. Kumar.
“Scalable Visualization and Interactive Analysis Using Massive Data Streams,” In Cloud Computing and Big Data, Advances in Parallel Computing, Vol. 23, IOS Press, pp. 212--230. 2013.
Historically, data creation and storage has always outpaced the infrastructure for its movement and utilization. This trend is increasing now more than ever, with the ever growing size of scientific simulations, increased resolution of sensors, and large mosaic images. Effective exploration of massive scientific models demands the combination of data management, analysis, and visualization techniques, working together in an interactive setting. The ViSUS application framework has been designed as an environment that allows the interactive exploration and analysis of massive scientific models in a cache-oblivious, hardware-agnostic manner, enabling processing and visualization of possibly geographically distributed data using many kinds of devices and platforms.
For general purpose feature segmentation and exploration we discuss a new paradigm based on topological analysis. This approach enables the extraction of summaries of features present in the data through abstract models that are orders of magnitude smaller than the raw data, providing enough information to support general queries and perform a wide range of analyses without access to the original data.
Keywords: Visualization, data analysis, topological data analysis, Parallel I/O
2012


J.C. Bennett, H. Abbasi, P. Bremer, R.W. Grout, A. Gyulassy, T. Jin, S. Klasky, H. Kolla, M. Parashar, V. Pascucci, P. Pbay, D. Thompson, H. Yu, F. Zhang, J. Chen.
“Combining In-Situ and In-Transit Processing to Enable Extreme-Scale Scientific Analysis,” In ACM/IEEE International Conference for High Performance Computing, Networking, Storage, and Analysis (SC), Salt Lake City, Utah, U.S.A., November, 2012.
With the onset of extreme-scale computing, I/O constraints make it increasingly difficult for scientists to save a sufficient amount of raw simulation data to persistent storage. One potential solution is to change the data analysis pipeline from a post-process centric to a concurrent approach based on either in-situ or in-transit processing. In this context computations are considered in-situ if they utilize the primary compute resources, while in-transit processing refers to offloading computations to a set of secondary resources using asynchronous data transfers. In this paper we explore the design and implementation of three common analysis techniques typically performed on large-scale scientific simulations: topological analysis, descriptive statistics, and visualization. We summarize algorithmic developments, describe a resource scheduling system to coordinate the execution of various analysis workflows, and discuss our implementation using the DataSpaces and ADIOS frameworks that support efficient data movement between in-situ and in-transit computations. We demonstrate the efficiency of our lightweight, flexible framework by deploying it on the Jaguar XK6 to analyze data generated by S3D, a massively parallel turbulent combustion code. Our framework allows scientists dealing with the data deluge at extreme scale to perform analyses at increased temporal resolutions, mitigate I/O costs, and significantly improve the time to insight.


A. Gyulassy, V. Pascucci, T. Peterka, R. Ross.
“The Parallel Computation of Morse-Smale Complexes,” In Proceedings of the Parallel and Distributed Processing Symposium (IPDPS), pp. 484--495. 2012.
DOI: 10.1109/IPDPS.2012.52
Topology-based techniques are useful for multiscale exploration of the feature space of scalar-valued functions, such as those derived from the output of large-scale simulations. The Morse-Smale (MS) complex, in particular, allows robust identification of gradient-based features, and therefore is suitable for analysis tasks in a wide range of application domains. In this paper, we develop a two-stage algorithm to construct the 1-skeleton of the Morse-Smale complex in parallel, the first stage independently computing local features per block and the second stage merging to resolve global features. Our implementation is based on MPI and a distributed-memory architecture. Through a set of scalability studies on the IBM Blue Gene/P supercomputer, we characterize the performance of the algorithm as block sizes, process counts, merging strategy, and levels of topological simplification are varied, for datasets that vary in feature composition and size. We conclude with a strong scaling study using scientific datasets computed by combustion and hydrodynamics simulations.


A. Gyulassy, P.-T. Bremer, V. Pascucci.
“Computing Morse-Smale Complexes with Accurate Geometry,” In IEEE Transactions on Visualization and Computer Graphics, Vol. 18, No. 12, pp. 2014--2022. 2012.
DOI: 10.1109/TVCG.2011.272
Topological techniques have proven highly successful in analyzing and visualizing scientific data. As a result, significant efforts have been made to compute structures like the Morse-Smale complex as robustly and efficiently as possible. However, the resulting algorithms, while topologically consistent, often produce incorrect connectivity as well as poor geometry. These problems may compromise or even invalidate any subsequent analysis. Moreover, such techniques may fail to improve even when the resolution of the domain mesh is increased, thus producing potentially incorrect results even for highly resolved functions. To address these problems we introduce two new algorithms: (i) a randomized algorithm to compute the discrete gradient of a scalar field that converges under refinement; and (ii) a deterministic variant which directly computes accurate geometry and thus correct connectivity of the MS complex. The first algorithm converges in the sense that on average it produces the correct result and its standard deviation approaches zero with increasing mesh resolution. The second algorithm uses two ordered traversals of the function to integrate the probabilities of the first to extract correct (near optimal) geometry and connectivity. We present an extensive empirical study using both synthetic and real-world data and demonstrates the advantages of our algorithms in comparison with several popular approaches.


A. Gyulassy, N. Kotava, M. Kim, C. Hansen, H. Hagen, and V. Pascucci.
“Direct Feature Visualization Using Morse-Smale Complexes,” In IEEE Transactions on Visualization and Computer Graphics, Vol. 18, No. 9, pp. 1549--1562. September, 2012.
DOI: 10.1109/TVCG.2011.272
In this paper, we characterize the range of features that can be extracted from an Morse-Smale complex and describe a unified query language to extract them. We provide a visual dictionary to guide users when defining features in terms of these queries. We demonstrate our topology-rich visualization pipeline in a tool that interactively queries the MS complex to extract features at multiple resolutions, assigns rendering attributes, and combines traditional volume visualization with the extracted features. The flexibility and power of this approach is illustrated with examples showing novel features.