Scientific Computing
Numerical simulation of real-world phenomena provides fertile ground for building interdisciplinary relationships. The SCI Institute has a long tradition of building these relationships in a win-win fashion – a win for the theoretical and algorithmic development of numerical modeling and simulation techniques and a win for the discipline-specific science of interest. High-order and adaptive methods, uncertainty quantification, complexity analysis, and parallelization are just some of the topics being investigated by SCI faculty. These areas of computing are being applied to a wide variety of engineering applications ranging from fluid mechanics and solid mechanics to bioelectricity.
Valerio Pascucci
Scientific Data Management
Chris Johnson
Problem Solving Environments
Ross Whitaker
GPUs
Chuck Hansen
GPUsFunded Research Projects:
Publications in Scientific Computing:
  Data and Range-Bounded Polynomials in ENO Methods M. Berzins. In Journal of Computational Science, Vol. 4, No. 1-2, pp. 62--70. 2013. DOI: 10.1016/j.jocs.2012.04.006 Essentially Non-Oscillatory (ENO) methods and Weighted Essentially Non-Oscillatory (WENO) methods are of fundamental importance in the numerical solution of hyperbolic equations. A key property of such equations is that the solution must remain positive or lie between bounds. A modification of the polynomials used in ENO methods to ensure that the modified polynomials are either bounded by adjacent values (data-bounded) or lie within a specified range (range-bounded) is considered. It is shown that this approach helps both in the range boundedness in the preservation of extrema in the ENO polynomial solution. |
  Effect of deltoid tension and humeral version in reverse total shoulder arthroplasty: a biomechanical study H.B. Henninger, Barg A, A.E. Anderson, K.N. Bachus, R.Z. Tashjian, R.T. Burks. In Journal of Shoulder and Elbow Surgery, Vol. 21, No. 4, pp. 483–-490. 2012. DOI: 10.1016/j.jse.2011.01.040 Background Materials and methods Results Conclusion Keywords: Shoulder, reverse arthroplasty, deltoid tension, humeral version, biomechanical simulator |
  Extreme-Scale Visual Analytics P.C. Wong, H. Shen, V. Pascucci. In IEEE Computer Graphics and Applications, Vol. 32, No. 4, pp. 23--25. 2012. DOI: 10.1109/MCG.2012.73 Extreme-scale visual analytics (VA) is about applying VA to extreme-scale data. The articles in this special issue examine advances related to extreme-scale VA problems, their analytical and computational challenges, and their real-world applications. |
  Atrial Fibrosis Quantified Using Late Gadolinium Enhancement MRI is AssociatedWith Sinus Node Dysfunction Requiring Pacemaker Implant N.W. Akoum, C.J. McGann, G. Vergara, T. Badger, R. Ranjan, C. Mahnkopf, E.G. Kholmovski, R.S. Macleod, N.F. Marrouche. In Journal of Cardiovascular Electrophysiology, Vol. 23, No. 1, pp. 44--50. 2012. DOI: 10.1111/j.1540-8167.2011.02140.x Atrial Fibrosis and Sinus Node Dysfunction. Introduction: Sinus node dysfunction (SND) commonly manifests with atrial arrhythmias alternating with sinus pauses and sinus bradycardia. The underlying process is thought to be because of atrial fibrosis. We assessed the value of atrial fibrosis, quantified using Late Gadolinium Enhanced-MRI (LGE-MRI), in predicting significant SND requiring pacemaker implant. Methods: Three hundred forty-four patients with atrial fibrillation (AF) presenting for catheter ablation underwent LGE-MRI. Left atrial (LA) fibrosis was quantified in all patients and right atrial (RA) fibrosis in 134 patients. All patients underwent catheter ablation with pulmonary vein isolation with posterior wall and septal debulking. Patients were followed prospectively for 329 ± 245 days. Ambulatory monitoring was instituted every 3 months. Symptomatic pauses and bradycardia were treated with pacemaker implantation per published guidelines. Results: The average patient age was 65 ± 12 years. The average wall fibrosis was 16.7 ± 11.1% in the LA, and 5.3 ± 6.4% in the RA. RA fibrosis was correlated with LA fibrosis (R2= 0.26; P < 0.01). Patients were divided into 4 stages of LA fibrosis (Utah I: 35%). Twenty-two patients (mean atrial fibrosis, 23.9%) required pacemaker implantation during follow-up. Univariate and multivariate analysis identified LA fibrosis stage (OR, 2.2) as a significant predictor for pacemaker implantation with an area under the curve of 0.704. Conclusions: In patients with AF presenting for catheter ablation, LGE-MRI quantification of atrial fibrosis demonstrates preferential LA involvement. Significant atrial fibrosis is associated with clinically significant SND requiring pacemaker implantation. (J Cardiovasc Electrophysiol, Vol. 23, pp. 44-50, January 2012) |
  30 Generation of Cloned Transgenic Goats with Cardiac Specific Overexpression of Transforming Growth Factor β1 Q. Meng, J. Hall, H. Rutigliano, X. Zhou, B.R. Sessions, R. Stott, K. Panter, C.J. Davies, R. Ranjan, D. Dosdall, R.S. MacLeod, N. Marrouche, K.L. White, Z. Wang, I.A. Polejaeva. In Reproduction, Fertility and Development, Vol. 25, No. 1, pp. 162--163. 2012. DOI: 10.1071/RDv25n1Ab30 Transforming growth factor β1 (TGF-β1) has a potent profibrotic function and is central to signaling cascades involved in interstitial fibrosis, which plays a critical role in the pathobiology of cardiomyopathy and contributes to diastolic and systolic dysfunction. In addition, fibrotic remodeling is responsible for generation of re-entry circuits that promote arrhythmias (Bujak and Frangogiannis 2007 Cardiovasc. Res. 74, 184–195). Due to the small size of the heart, functional electrophysiology of transgenic mice is problematic. Large transgenic animal models have the potential to offer insights into conduction heterogeneity associated with fibrosis and the role of fibrosis in cardiovascular diseases. The goal of this study was to generate transgenic goats overexpressing an active form of TGFβ-1 under control of the cardiac-specific α-myosin heavy chain promoter (α-MHC). A pcDNA3.1DV5-MHC-TGF-β1cys33ser vector was constructed by subcloning the MHC-TGF-β1 fragment from the plasmid pUC-BM20-MHC-TGF-β1 (Nakajima et al. 2000 Circ. Res. 86, 571–579) into the pcDNA3.1D V5 vector. The Neon transfection system was used to electroporate primary goat fetal fibroblasts. After G418 selection and PCR screening, transgenic cells were used for SCNT. Oocytes were collected by slicing ovaries from an abattoir and matured in vitro in an incubator with 5\% CO2 in air. Cumulus cells were removed at 21 to 23 h post-maturation. Oocytes were enucleated by aspirating the first polar body and nearby cytoplasm by micromanipulation in Hepes-buffered SOF medium with 10 µg of cytochalasin B mL–1. Transgenic somatic cells were individually inserted into the perivitelline space and fused with enucleated oocytes using double electrical pulses of 1.8 kV cm–1 (40 µs each). Reconstructed embryos were activated by ionomycin (5 min) and DMAP and cycloheximide (CHX) treatments. Cloned embryos were cultured in G1 medium for 12 to 60 h in vitro and then transferred into synchronized recipient females. Pregnancy was examined by ultrasonography on day 30 post-transfer. A total of 246 cloned embryos were transferred into 14 recipients that resulted in production of 7 kids. The pregnancy rate was higher in the group cultured for 12 h compared with those cultured 36 to 60 h [44.4\% (n = 9) v. 20\% (n = 5)]. The kidding rates per embryo transferred of these 2 groups were 3.8\% (n = 156) and 1.1\% (n = 90), respectively. The PCR results confirmed that all the clones were transgenic. Phenotype characterization [e.g. gene expression, electrocardiogram (ECG), and magnetic resonance imaging (MRI)] is underway. We demonstrated successful production of transgenic goat via SCNT. To our knowledge, this is the first transgenic goat model produced for cardiovascular research. |
  Stochastic Collocation for Optimal Control Problems with Stochastic PDE Constraints H. Tiesler, R.M. Kirby, D. Xiu, T. Preusser. In SIAM Journal on Control and Optimization, Vol. 50, No. 5, pp. 2659--2682. 2012. DOI: 10.1137/110835438 We discuss the use of stochastic collocation for the solution of optimal control problems which are constrained by stochastic partial differential equations (SPDE). Thereby the constraining, SPDE depends on data which is not deterministic but random. Assuming a deterministic control, randomness within the states of the input data will propagate to the states of the system. For the solution of SPDEs there has recently been an increasing effort in the development of efficient numerical schemes based upon the mathematical concept of generalized polynomial chaos. Modal-based stochastic Galerkin and nodal-based stochastic collocation versions of this methodology exist, both of which rely on a certain level of smoothness of the solution in the random space to yield accelerated convergence rates. In this paper we apply the stochastic collocation method to develop a gradient descent as well as a sequential quadratic program (SQP) for the minimization of objective functions constrained by an SPDE. The stochastic function involves several higher-order moments of the random states of the system as well as classical regularization of the control. In particular we discuss several objective functions of tracking type. Numerical examples are presented to demonstrate the performance of our new stochastic collocation minimization approach. Keywords: stochastic collocation, optimal control, stochastic partial differential equations |
  Efficient data restructuring and aggregation for I/O acceleration in PIDX S. Kumar, V. Vishwanath, P. Carns, J.A. Levine, R. Latham, G. Scorzelli, H. Kolla, R. Grout, R. Ross, M.E. Papka, J. Chen, V. Pascucci. In Proceedings of the International Conference on High Performance Computing, Networking, Storage and Analysis, IEEE Computer Society Press, pp. 50:1--50:11. 2012. ISBN: 978-1-4673-0804-5 Hierarchical, multiresolution data representations enable interactive analysis and visualization of large-scale simulations. One promising application of these techniques is to store high performance computing simulation output in a hierarchical Z (HZ) ordering that translates data from a Cartesian coordinate scheme to a one-dimensional array ordered by locality at different resolution levels. However, when the dimensions of the simulation data are not an even power of 2, parallel HZ ordering produces sparse memory and network access patterns that inhibit I/O performance. This work presents a new technique for parallel HZ ordering of simulation datasets that restructures simulation data into large (power of 2) blocks to facilitate efficient I/O aggregation. We perform both weak and strong scaling experiments using the S3D combustion application on both Cray-XE6 (65,536 cores) and IBM Blue Gene/P (131,072 cores) platforms. We demonstrate that data can be written in hierarchical, multiresolution format with performance competitive to that of native data-ordering methods. |
  Smoothness-Increasing Accuracy-Conserving (SIAC) Filtering for discontinuous Galerkin Solutions: Improved Errors Versus Higher-Order Accuracy J. King, H. Mirzaee, J.K. Ryan, R.M. Kirby. In Journal of Scientific Computing, Vol. 53, pp. 129--149. 2012. DOI: 10.1007/s10915-012-9593-8 Smoothness-increasing accuracy-conserving (SIAC) filtering has demonstrated its effectiveness in raising the convergence rate of discontinuous Galerkin solutions from order k + 1/2 to order 2k + 1 for specific types of translation invariant meshes (Cockburn et al. in Math. Comput. 72:577–606, 2003; Curtis et al. in SIAM J. Sci. Comput. 30(1):272– 289, 2007; Mirzaee et al. in SIAM J. Numer. Anal. 49:1899–1920, 2011). Additionally, it improves the weak continuity in the discontinuous Galerkin method to k - 1 continuity. Typically this improvement has a positive impact on the error quantity in the sense that it also reduces the absolute errors. However, not enough emphasis has been placed on the difference between superconvergent accuracy and improved errors. This distinction is particularly important when it comes to understanding the interplay introduced through meshing, between geometry and filtering. The underlying mesh over which the DG solution is built is important because the tool used in SIAC filtering—convolution—is scaled by the geometric mesh size. This heavily contributes to the effectiveness of the post-processor. In this paper, we present a study of this mesh scaling and how it factors into the theoretical errors. To accomplish the large volume of post-processing necessary for this study, commodity streaming multiprocessors were used; we demonstrate for structured meshes up to a 50× speed up in the computational time over traditional CPU implementations of the SIAC filter. |
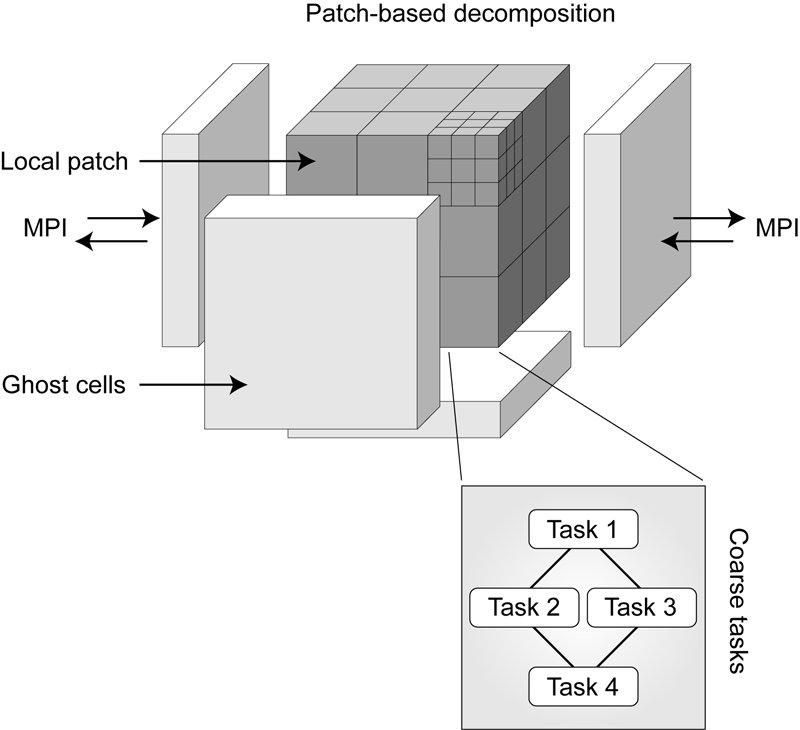
  Multiscale Modeling of High Explosives for Transportation Accidents J.R. Peterson, J.C. Beckvermit, T. Harman, M. Berzins, C.A. Wight. In Proceedings of the 1st Conference of the Extreme Science and Engineering Discovery Environment: Bridging from the eXtreme to the campus and beyond, 2012. DOI: 10.1145/2335755.2335828 The development of a reaction model to simulate the accidental detonation of a large array of seismic boosters in a semi-truck subject to fire is considered. To test this model large scale simulations of explosions and detonations were performed by leveraging the massively parallel capabilities of the Uintah Computational Framework and the XSEDE computational resources. Computed stress profiles in bulk-scale explosive materials were validated using compaction simulations of hundred micron scale particles and found to compare favorably with experimental data. A validation study of reaction models for deflagration and detonation showed that computational grid cell sizes up to 10 mm could be used without loss of fidelity. The Uintah Computational Framework shows linear scaling up to 180K cores which combined with coarse resolution and validated models will now enable simulations of semi-truck scale transportation accidents for the first time. |
  Radiation Modeling Using the Uintah Heterogeneous CPU/GPU Runtime System A. Humphrey, Q. Meng, M. Berzins, T. Harman. In Proceedings of the first conference of the Extreme Science and Engineering Discovery Environment (XSEDE'12), Association for Computing Machinery, 2012. DOI: 10.1145/2335755.2335791 The Uintah Computational Framework was developed to provide an environment for solving fluid-structure interaction problems on structured adaptive grids on large-scale, long-running, data-intensive problems. Uintah uses a combination of fluid-flow solvers and particle-based methods for solids, together with a novel asynchronous task-based approach with fully automated load balancing. Uintah demonstrates excellent weak and strong scalability at full machine capacity on XSEDE resources such as Ranger and Kraken, and through the use of a hybrid memory approach based on a combination of MPI and Pthreads, Uintah now runs on up to 262k cores on the DOE Jaguar system. In order to extend Uintah to heterogeneous systems, with ever-increasing CPU core counts and additional onnode GPUs, a new dynamic CPU-GPU task scheduler is designed and evaluated in this study. This new scheduler enables Uintah to fully exploit these architectures with support for asynchronous, outof- order scheduling of both CPU and GPU computational tasks. A new runtime system has also been implemented with an added multi-stage queuing architecture for efficient scheduling of CPU and GPU tasks. This new runtime system automatically handles the details of asynchronous memory copies to and from the GPU and introduces a novel method of pre-fetching and preparing GPU memory prior to GPU task execution. In this study this new design is examined in the context of a developing, hierarchical GPUbased ray tracing radiation transport model that provides Uintah with additional capabilities for heat transfer and electromagnetic wave propagation. The capabilities of this new scheduler design are tested by running at large scale on the modern heterogeneous systems, Keeneland and TitanDev, with up to 360 and 960 GPUs respectively. On these systems, we demonstrate significant speedups per GPU against a standard CPU core for our radiation problem. Keywords: Uintah, hybrid parallelism, scalability, parallel, adaptive, GPU, heterogeneous systems, Keeneland, TitanDev |
  Biomedical Visual Computing: Case Studies and Challenges C.R. Johnson. In IEEE Computing in Science and Engineering, Vol. 14, No. 1, pp. 12--21. 2012. PubMed ID: 22545005 PubMed Central ID: PMC3336198 Computer simulation and visualization are having a substantial impact on biomedicine and other areas of science and engineering. Advanced simulation and data acquisition techniques allow biomedical researchers to investigate increasingly sophisticated biological function and structure. A continuing trend in all computational science and engineering applications is the increasing size of resulting datasets. This trend is also evident in data acquisition, especially in image acquisition in biology and medical image databases. For example, in a collaboration between neuroscientist Robert Marc and our research team at the University of Utah's Scientific Computing and Imaging (SCI) Institute (www.sci.utah.edu), we're creating datasets of brain electron microscopy (EM) mosaics that are 16 terabytes in size. However, while there's no foreseeable end to the increase in our ability to produce simulation data or record observational data, our ability to use this data in meaningful ways is inhibited by current data analysis capabilities, which already lag far behind. Indeed, as the NIH-NSF Visualization Research Challenges report notes, to effectively understand and make use of the vast amounts of data researchers are producing is one of the greatest scientific challenges of the 21st century. Visual data analysis involves creating images that convey salient information about underlying data and processes, enabling the detection and validation of expected results while leading to unexpected discoveries in science. This allows for the validation of new theoretical models, provides comparison between models and datasets, enables quantitative and qualitative querying, improves interpretation of data, and facilitates decision making. Scientists can use visual data analysis systems to explore \"what if\" scenarios, define hypotheses, and examine data under multiple perspectives and assumptions. In addition, they can identify connections between numerous attributes and quantitatively assess the reliability of hypotheses. In essence, visual data analysis is an integral part of scientific problem solving and discovery. As applied to biomedical systems, visualization plays a crucial role in our ability to comprehend large and complex data-data that, in two, three, or more dimensions, convey insight into many diverse biomedical applications, including understanding neural connectivity within the brain, interpreting bioelectric currents within the heart, characterizing white-matter tracts by diffusion tensor imaging, and understanding morphology differences among different genetic mice phenotypes. Keywords: kaust |
  Fast, Effective BVH Updates for Animated Scenes D. Kopta, T. Ize, J. Spjut, E. Brunvand, A. Davis, A. Kensler. In Proceedings of the Symposium on Interactive 3D Graphics and Games (I3D '12), pp. 197--204. 2012. DOI: 10.1145/2159616.2159649 Bounding volume hierarchies (BVHs) are a popular acceleration structure choice for animated scenes rendered with ray tracing. This is due to the relative simplicity of refitting bounding volumes around moving geometry. However, the quality of such a refitted tree can degrade rapidly if objects in the scene deform or rearrange significantly as the animation progresses, resulting in dramatic increases in rendering times and a commensurate reduction in the frame rate. The BVH could be rebuilt on every frame, but this could take significant time. We present a method to efficiently extend refitting for animated scenes with tree rotations, a technique previously proposed for off-line improvement of BVH quality for static scenes. Tree rotations are local restructuring operations which can mitigate the effects that moving primitives have on BVH quality by rearranging nodes in the tree during each refit rather than triggering a full rebuild. The result is a fast, lightweight, incremental update algorithm that requires negligible memory, has minor update times, parallelizes easily, avoids significant degradation in tree quality or the need for rebuilding, and maintains fast rendering times. We show that our method approaches or exceeds the frame rates of other techniques and is consistently among the best options regardless of the animated scene. |