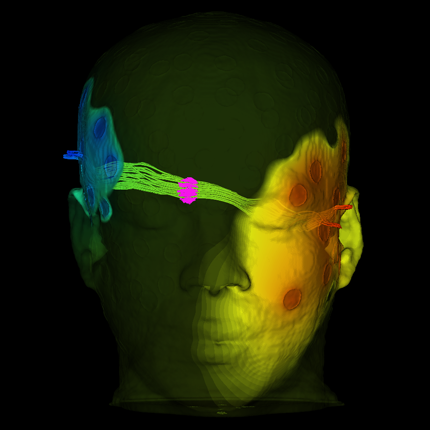
Visualization
Visualization, sometimes referred to as visual data analysis, uses the graphical representation of data as a means of gaining understanding and insight into the data. Visualization research at SCI has focused on applications spanning computational fluid dynamics, medical imaging and analysis, biomedical data analysis, healthcare data analysis, weather data analysis, poetry, network and graph analysis, financial data analysis, etc.Research involves novel algorithm and technique development to building tools and systems that assist in the comprehension of massive amounts of (scientific) data. We also research the process of creating successful visualizations.
We strongly believe in the role of interactivity in visual data analysis. Therefore, much of our research is concerned with creating visualizations that are intuitive to interact with and also render at interactive rates.
Visualization at SCI includes the academic subfields of Scientific Visualization, Information Visualization and Visual Analytics.

Mike Kirby
Uncertainty Visualization
Alex Lex
Information VisualizationCenters and Labs:
- Visualization Design Lab (VDL)
- CEDMAV
- POWDER Display Wall
- Modeling, Display, and Understanding Uncertainty in Simulations for Policy Decision Making
- Topological Data Analysis for Large Network Visualization
Funded Research Projects:
Publications in Visualization:
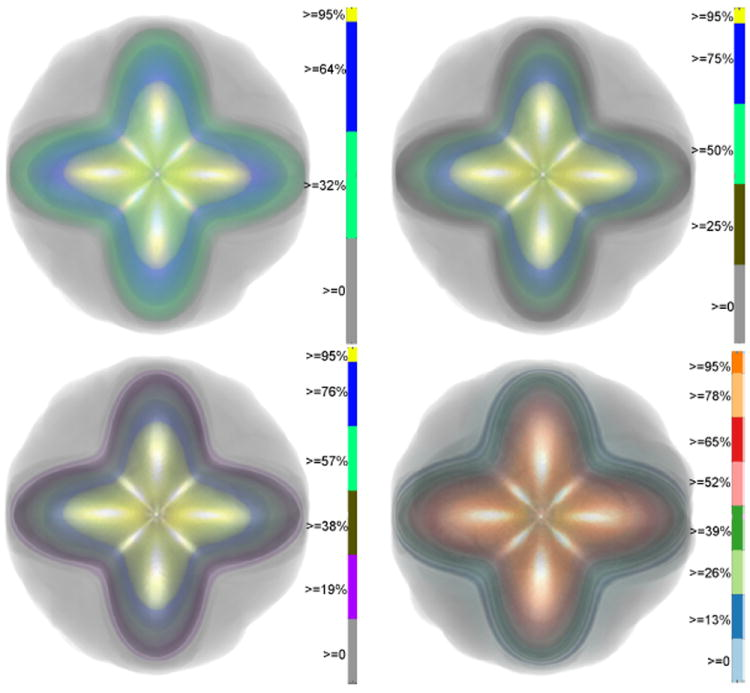
  Uncertainty Visualization in HARDI based on Ensembles of ODFs F. Jiao, J.M. Phillips, Y. Gur, C.R. Johnson. In Proceedings of 2013 IEEE Pacific Visualization Symposium, pp. 193--200. 2013. PubMed ID: 24466504 PubMed Central ID: PMC3898522 In this paper, we propose a new and accurate technique for uncertainty analysis and uncertainty visualization based on fiber orientation distribution function (ODF) glyphs, associated with high angular resolution diffusion imaging (HARDI). Our visualization applies volume rendering techniques to an ensemble of 3D ODF glyphs, which we call SIP functions of diffusion shapes, to capture their variability due to underlying uncertainty. This rendering elucidates the complex heteroscedastic structural variation in these shapes. Furthermore, we quantify the extent of this variation by measuring the fraction of the volume of these shapes, which is consistent across all noise levels, the certain volume ratio. Our uncertainty analysis and visualization framework is then applied to synthetic data, as well as to HARDI human-brain data, to study the impact of various image acquisition parameters and background noise levels on the diffusion shapes. |
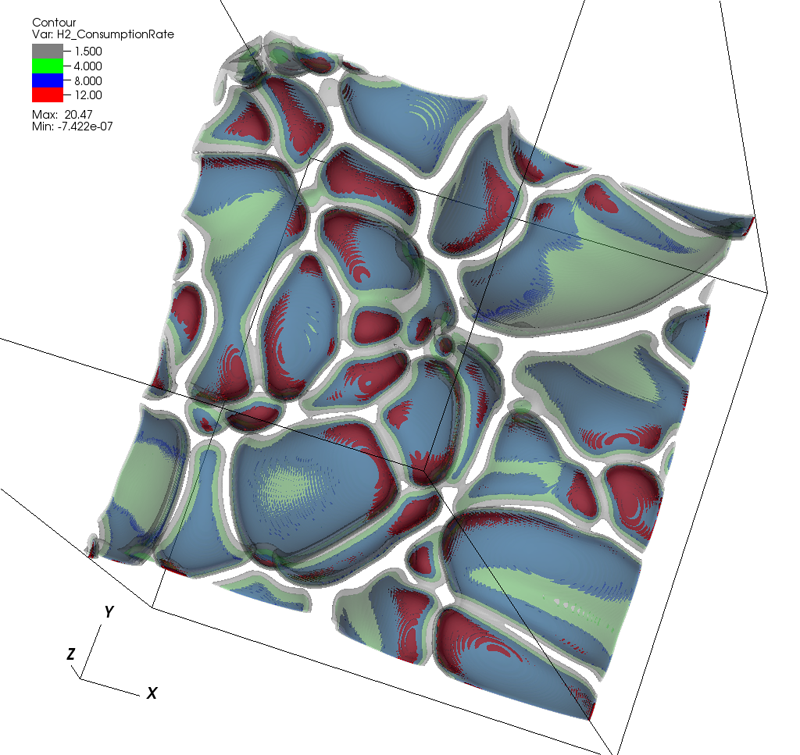
  Topology analysis of time-dependent multi-fluid data using the Reeb graph F. Chen, H. Obermaier, H. Hagen, B. Hamann, J. Tierny, V. Pascucci. In Computer Aided Geometric Design, Vol. 30, No. 6, pp. 557--566. 2013. DOI: 10.1016/j.cagd.2012.03.019 Liquid–liquid extraction is a typical multi-fluid problem in chemical engineering where two types of immiscible fluids are mixed together. Mixing of two-phase fluids results in a time-varying fluid density distribution, quantitatively indicating the presence of liquid phases. For engineers who design extraction devices, it is crucial to understand the density distribution of each fluid, particularly flow regions that have a high concentration of the dispersed phase. The propagation of regions of high density can be studied by examining the topology of isosurfaces of the density data. We present a topology-based approach to track the splitting and merging events of these regions using the Reeb graphs. Time is used as the third dimension in addition to two-dimensional (2D) point-based simulation data. Due to low time resolution of the input data set, a physics-based interpolation scheme is required in order to improve the accuracy of the proposed topology tracking method. The model used for interpolation produces a smooth time-dependent density field by applying Lagrangian-based advection to the given simulated point cloud data, conforming to the physical laws of flow evolution. Using the Reeb graph, the spatial and temporal locations of bifurcation and merging events can be readily identified supporting in-depth analysis of the extraction process. Keywords: Multi-phase fluid, Level set, Topology method, Point-based multi-fluid simulation |
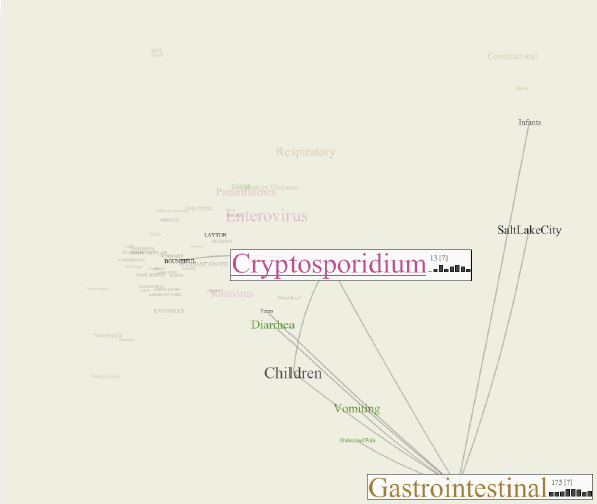
  The CommonGround visual paradigm for biosurveillance Y. Livnat, E. Jurrus, A.V. Gundlapalli, P. Gestland. In Proceedings of the 2013 IEEE International Conference on Intelligence and Security Informatics (ISI), pp. 352--357. 2013. ISBN: 978-1-4673-6214-6 DOI: 10.1109/ISI.2013.6578857 Biosurveillance is a critical area in the intelligence community for real-time detection of disease outbreaks. Identifying epidemics enables analysts to detect and monitor disease outbreak that might be spread from natural causes or from possible biological warfare attacks. Containing these events and disseminating alerts requires the ability to rapidly find, classify and track harmful biological signatures. In this paper, we describe a novel visual paradigm to conduct biosurveillance using an Infectious Disease Weather Map. Our system provides a visual common ground in which users can view, explore and discover emerging concepts and correlations such as symptoms, syndromes, pathogens and geographic locations. Keywords: biosurveillance, visualization, interactive exploration, situational awareness |
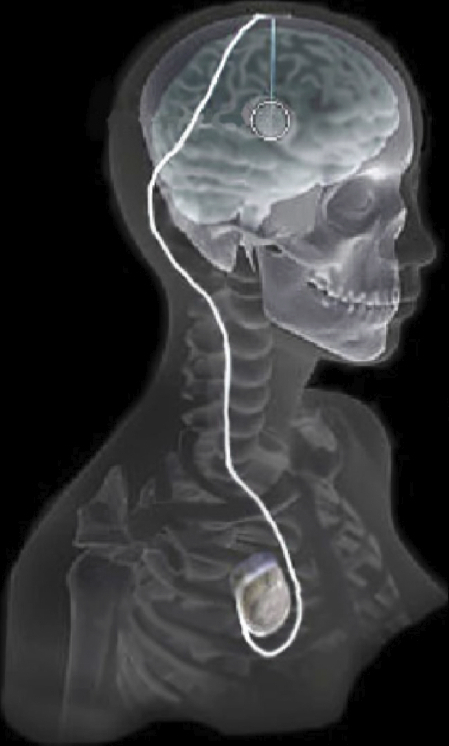
  Evaluation of Interactive Visualization on Mobile Computing Platforms for Selection of Deep Brain Stimulation Parameters C. Butson, G. Tamm, S. Jain, T. Fogal, J. Krüger. In IEEE Transactions on Visualization and Computer Graphics, Vol. 19, No. 1, pp. 108--117. January, 2013. DOI: 10.1109/TVCG.2012.92 PubMed ID: 22450824 In recent years there has been significant growth in the use of patient-specific models to predict the effects of neuromodulation therapies such as deep brain stimulation (DBS). However, translating these models from a research environment to the everyday clinical workflow has been a challenge, primarily due to the complexity of the models and the expertise required in specialized visualization software. In this paper, we deploy the interactive visualization system ImageVis3D Mobile , which has been designed for mobile computing devices such as the iPhone or iPad, in an evaluation environment to visualize models of Parkinson’s disease patients who received DBS therapy. Selection of DBS settings is a significant clinical challenge that requires repeated revisions to achieve optimal therapeutic response, and is often performed without any visual representation of the stimulation system in the patient. We used ImageVis3D Mobile to provide models to movement disorders clinicians and asked them to use the software to determine: 1) which of the four DBS electrode contacts they would select for therapy; and 2) what stimulation settings they would choose. We compared the stimulation protocol chosen from the software versus the stimulation protocol that was chosen via clinical practice (independently of the study). Lastly, we compared the amount of time required to reach these settings using the software versus the time required through standard practice. We found that the stimulation settings chosen using ImageVis3D Mobile were similar to those used in standard of care, but were selected in drastically less time. We show how our visualization system, available directly at the point of care on a device familiar to the clinician, can be used to guide clinical decision making for selection of DBS settings. In our view, the positive impact of the system could also translate to areas other than DBS. Keywords: Biomedical and Medical Visualization, Mobile and Ubiquitous Visualization, Computational Model, Clinical Decision Making, Parkinson’s Disease, SciDAC, ImageVis3D |
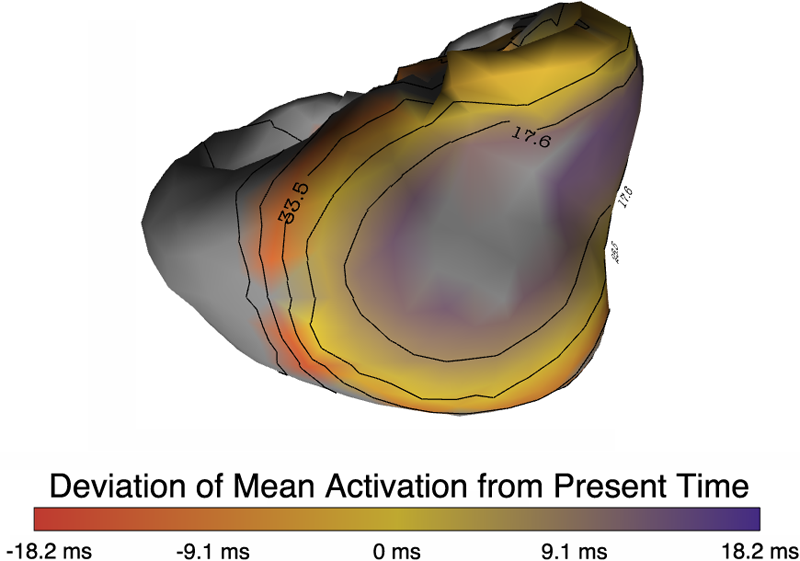
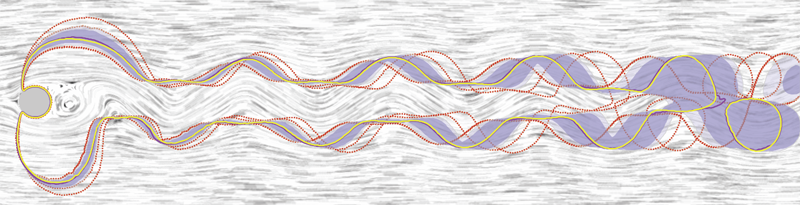
  Contour Boxplots: A Method for Characterizing Uncertainty in Feature Sets from Simulation Ensembles R.T. Whitaker, M. Mirzargar, R.M. Kirby. In IEEE Transactions on Visualization and Computer Graphics, Vol. 19, No. 12, pp. 2713--2722. December, 2013. DOI: 10.1109/TVCG.2013.143 PubMed ID: 24051838 Ensembles of numerical simulations are used in a variety of applications, such as meteorology or computational solid mechanics, in order to quantify the uncertainty or possible error in a model or simulation. Deriving robust statistics and visualizing the variability of an ensemble is a challenging task and is usually accomplished through direct visualization of ensemble members or by providing aggregate representations such as an average or pointwise probabilities. In many cases, the interesting quantities in a simulation are not dense fields, but are sets of features that are often represented as thresholds on physical or derived quantities. In this paper, we introduce a generalization of boxplots, called contour boxplots, for visualization and exploration of ensembles of contours or level sets of functions. Conventional boxplots have been widely used as an exploratory or communicative tool for data analysis, and they typically show the median, mean, confidence intervals, and outliers of a population. The proposed contour boxplots are a generalization of functional boxplots, which build on the notion of data depth. Data depth approximates the extent to which a particular sample is centrally located within its density function. This produces a center-outward ordering that gives rise to the statistical quantities that are essential to boxplots. Here we present a generalization of functional data depth to contours and demonstrate methods for displaying the resulting boxplots for two-dimensional simulation data in weather forecasting and computational fluid dynamics. |
  Adaptive Sampling with Topological Scores D. Maljovec, Bei Wang, A. Kupresanin, G. Johannesson, V. Pascucci, P.-T. Bremer. In Int. J. Uncertainty Quantification, Vol. 3, No. 2, Begell House, pp. 119--141. 2013. DOI: 10.1615/int.j.uncertaintyquantification.2012003955 Understanding and describing expensive black box functions such as physical simulations is a common problem in many application areas. One example is the recent interest in uncertainty quantification with the goal of discovering the relationship between a potentially large number of input parameters and the output of a simulation. Typically, the simulation of interest is expensive to evaluate and thus the sampling of the parameter space is necessarily small. As a result choosing a "good" set of samples at which to evaluate is crucial to glean as much information as possible from the fewest samples. While space-filling sampling designs such as Latin hypercubes provide a good initial cover of the entire domain, more detailed studies typically rely on adaptive sampling: Given an initial set of samples, these techniques construct a surrogate model and use it to evaluate a scoring function which aims to predict the expected gain from evaluating a potential new sample. There exist a large number of different surrogate models as well as different scoring functions each with their own advantages and disadvantages. In this paper we present an extensive comparative study of adaptive sampling using four popular regression models combined with six traditional scoring functions compared against a space-filling design. Furthermore, for a single high-dimensional output function, we introduce a new class of scoring functions based on global topological rather than local geometric information. The new scoring functions are competitive in terms of the root mean squared prediction error but are expected to better recover the global topological structure. Our experiments suggest that the most common point of failure of adaptive sampling schemes are ill-suited regression models. Nevertheless, even given well-fitted surrogate models many scoring functions fail to outperform a space-filling design. |
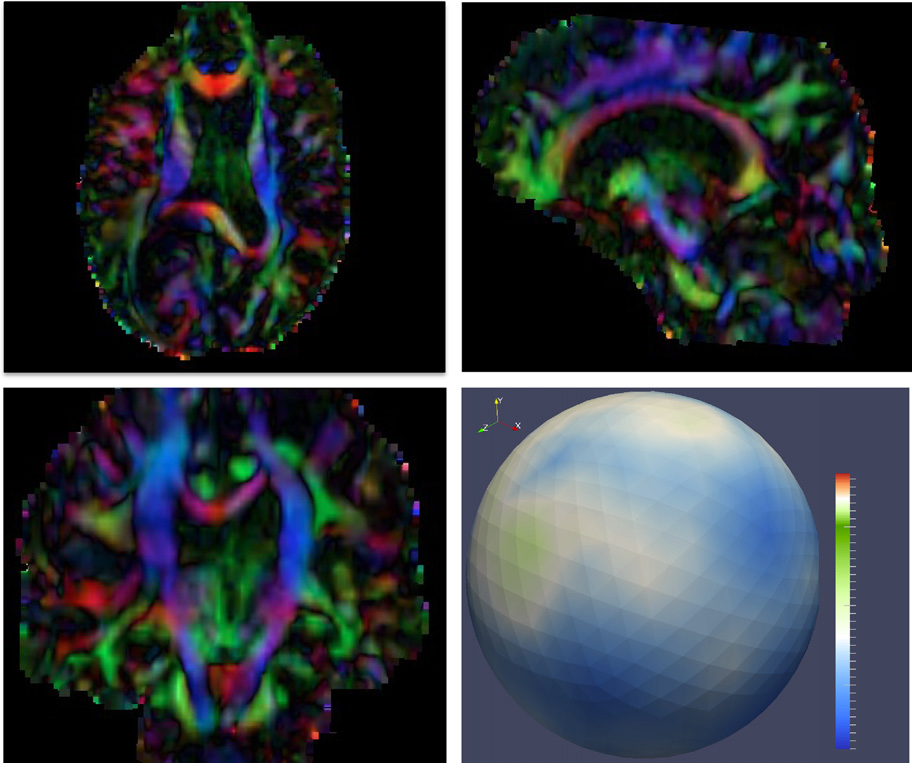
  Diffusion imaging quality control via entropy of principal direction distribution, M. Farzinfar, I. Oguz, R.G. Smith, A.R. Verde, C. Dietrich, A. Gupta, M.L. Escolar, J. Piven, S. Pujol, C. Vachet, S. Gouttard, G. Gerig, S. Dager, R.C. McKinstry, S. Paterson, A.C. Evans, M.A. Styner. In NeuroImage, Vol. 82, pp. 1--12. 2013. ISSN: 1053-8119 DOI: 10.1016/j.neuroimage.2013.05.022 Diffusion MR imaging has received increasing attention in the neuroimaging community, as it yields new insights into the microstructural organization of white matter that are not available with conventional MRI techniques. While the technology has enormous potential, diffusion MRI suffers from a unique and complex set of image quality problems, limiting the sensitivity of studies and reducing the accuracy of findings. Furthermore, the acquisition time for diffusion MRI is longer than conventional MRI due to the need for multiple acquisitions to obtain directionally encoded Diffusion Weighted Images (DWI). This leads to increased motion artifacts, reduced signal-to-noise ratio (SNR), and increased proneness to a wide variety of artifacts, including eddy-current and motion artifacts, “venetian blind” artifacts, as well as slice-wise and gradient-wise inconsistencies. Such artifacts mandate stringent Quality Control (QC) schemes in the processing of diffusion MRI data. Most existing QC procedures are conducted in the DWI domain and/or on a voxel level, but our own experiments show that these methods often do not fully detect and eliminate certain types of artifacts, often only visible when investigating groups of DWI's or a derived diffusion model, such as the most-employed diffusion tensor imaging (DTI). Here, we propose a novel regional QC measure in the DTI domain that employs the entropy of the regional distribution of the principal directions (PD). The PD entropy quantifies the scattering and spread of the principal diffusion directions and is invariant to the patient's position in the scanner. High entropy value indicates that the PDs are distributed relatively uniformly, while low entropy value indicates the presence of clusters in the PD distribution. The novel QC measure is intended to complement the existing set of QC procedures by detecting and correcting residual artifacts. Such residual artifacts cause directional bias in the measured PD and here called dominant direction artifacts. Experiments show that our automatic method can reliably detect and potentially correct such artifacts, especially the ones caused by the vibrations of the scanner table during the scan. The results further indicate the usefulness of this method for general quality assessment in DTI studies. Keywords: Diffusion magnetic resonance imaging, Diffusion tensor imaging, Quality assessment, Entropy |
  The Helmholtz-Hodge Decomposition - A Survey H. Bhatia, G. Norgard, V. Pascucci, P.-T. Bremer. In IEEE Transactions on Visualization and Computer Graphics (TVCG), Vol. 19, No. 8, Note: Selected as Spotlight paper for August 2013 issue, pp. 1386--1404. 2013. DOI: 10.1109/TVCG.2012.316 The Helmholtz-Hodge Decomposition (HHD) describes the decomposition of a flow field into its divergence-free and curl-free components. Many researchers in various communities like weather modeling, oceanology, geophysics, and computer graphics are interested in understanding the properties of flow representing physical phenomena such as incompressibility and vorticity. The HHD has proven to be an important tool in the analysis of fluids, making it one of the fundamental theorems in fluid dynamics. The recent advances in the area of flow analysis have led to the application of the HHD in a number of research communities such as flow visualization, topological analysis, imaging, and robotics. However, because the initial body of work, primarily in the physics communities, research on the topic has become fragmented with different communities working largely in isolation often repeating and sometimes contradicting each others results. |
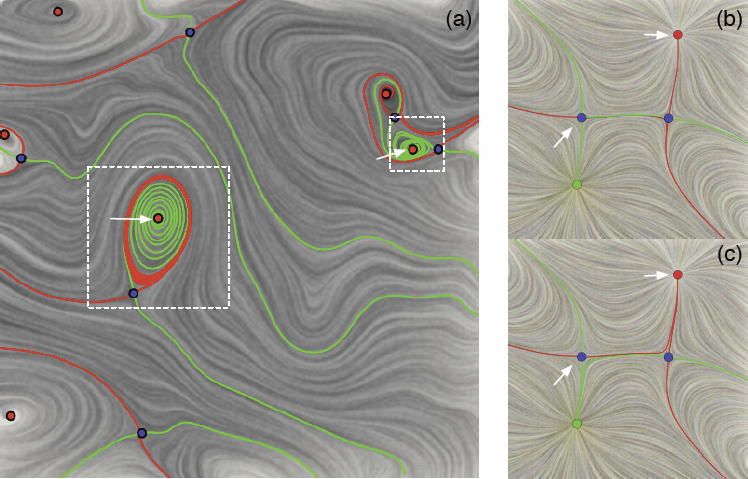
  Visualizing Robustness of Critical Points for 2D Time-Varying Vector Fields, Bei Wang, P. Rosen, P. Skraba, H. Bhatia, V. Pascucci. In Computer Graphics Forum, Vol. 32, No. 3, Wiley-Blackwell, pp. 221--230. jun, 2013. DOI: 10.1111/cgf.12109 Analyzing critical points and their temporal evolutions plays a crucial role in understanding the behavior of vector fields. A key challenge is to quantify the stability of critical points: more stable points may represent more important phenomena or vice versa. The topological notion of robustness is a tool which allows us to quantify rigorously the stability of each critical point. Intuitively, the robustness of a critical point is the minimum amount of perturbation necessary to cancel it within a local neighborhood, measured under an appropriate metric. In this paper, we introduce a new analysis and visualization framework which enables interactive exploration of robustness of critical points for both stationary and time-varying 2D vector fields. This framework allows the end-users, for the first time, to investigate how the stability of a critical point evolves over time. We show that this depends heavily on the global properties of the vector field and that structural changes can correspond to interesting behavior. We demonstrate the practicality of our theories and techniques on several datasets involving combustion and oceanic eddy simulations and obtain some key insights regarding their stable and unstable features. |