Image Analysis
SCI's imaging work addresses fundamental questions in 2D and 3D image processing, including filtering, segmentation, surface reconstruction, and shape analysis. In low-level image processing, this effort has produce new nonparametric methods for modeling image statistics, which have resulted in better algorithms for denoising and reconstruction. Work with particle systems has led to new methods for visualizing and analyzing 3D surfaces. Our work in image processing also includes applications of advanced computing to 3D images, which has resulted in new parallel algorithms and real-time implementations on graphics processing units (GPUs). Application areas include medical image analysis, biological image processing, defense, environmental monitoring, and oil and gas.
Ross Whitaker
Segmentation
Chris Johnson
Diffusion Tensor AnalysisFunded Research Projects:
Publications in Image Analysis:
 Quantifying Impurity Effects on the Surface Morphology of α-U3O8, A. B. Hanson, R. N. Lee, C. Vachet, I. J. Schwerdt, T. Tasdizen, L. W. McDonald IV. In Analytical Chemistry, 2019. DOI: doi:10.1021/acs.analchem.9b02013 The morphological effect of impurities on α-U3O8 has been investigated. This study provides the first evidence that the presence of impurities can alter nuclear material morphology, and these changes can be quantified to aid in revealing processing history. Four elements: Ca, Mg, V, and Zr were implemented in the uranyl peroxide synthesis route and studied individually within the α-U3O8. Six total replicates were synthesized, and replicates 1–3 were filtered and washed with Millipore water (18.2 MΩ) to remove any residual nitrates. Replicates 4–6 were filtered but not washed to determine the amount of impurities removed during washing. Inductively coupled plasma mass spectrometry (ICP-MS) was employed at key points during the synthesis to quantify incorporation of the impurity. Each sample was characterized using powder X-ray diffraction (p-XRD), high-resolution scanning electron microscopy (HRSEM), and SEM with energy dispersive X-ray spectroscopy (SEM-EDS). p-XRD was utilized to evaluate any crystallographic changes due to the impurities; HRSEM imagery was analyzed with Morphological Analysis for MAterials (MAMA) software and machine learning classification for quantification of the morphology; and SEM-EDS was utilized to locate the impurity within the α-U3O8. All samples were found to be quantifiably distinguishable, further demonstrating the utility of quantitative morphology as a signature for the processing history of nuclear material. |
  Image-based analysis and long-term clinical outcomes of deep brain stimulation for Tourette syndrome: a multisite study K. A. Johnson, P. T. Fletcher, D. Servello, A. Bona, M. Porta, J. L. Ostrem, E. Bardinet, M. Welter, A. M. Lozano, J. C. Baldermann, J. Kuhn, D. Huys, T. Foltynie, M. Hariz, E. M. Joyce, L. Zrinzo, Z. Kefalopoulou, J. Zhang, F. Meng, C. Zhang, Z. Ling, X. Xu, X. Yu, A. YJM Smeets, L. Ackermans, V. Visser-Vandewalle, A. Y. Mogilner, M. H. Pourfar, L. Almeida, A. Gunduz, W. Hu, K. D. Foote, M. S. Okun, C. R. Butson. In Journal of Neurology, Neurosurgery & Psychiatry, BMJ Publishing Group, 2019. DOI: 10.1136/jnnp-2019-320379 BACKGROUND: METHODS:
We collected retrospective clinical data and imaging from 13 international sites on 123 patients. We assessed the effects of DBS over time in 110 patients who were implanted in the centromedial (CM) thalamus (n=51), globus pallidus internus (GPi) (n=47), nucleus accumbens/anterior limb of the internal capsule (n=4) or a combination of targets (n=8). Contact locations (n=70 patients) and volumes of tissue activated (n=63 patients) were coregistered to create probabilistic stimulation atlases.RESULTS:
Tics and obsessive-compulsive behaviour (OCB) significantly improved over time (p<0.01), and there were no significant differences across brain targets (p>0.05). The median time was 13 months to reach a 40% improvement in tics, and there were no significant differences across targets (p=0.84), presence of OCB (p=0.09) or age at implantation (p=0.08). Active contacts were generally clustered near the target nuclei, with some variability that may reflect differences in targeting protocols, lead models and contact configurations. There were regions within and surrounding GPi and CM thalamus that improved tics for some patients but were ineffective for others. Regions within, superior or medial to GPi were associated with a greater improvement in OCB than regions inferior to GPi.CONCLUSION:
The results collectively indicate that DBS may improve tics and OCB, the effects may develop over several months, and stimulation locations relative to structural anatomy alone may not predict response. This study was the first to visualise and evaluate the regions of stimulation across a large cohort of patients with TS to generate new hypotheses about potential targets for improving tics and comorbidities.
|
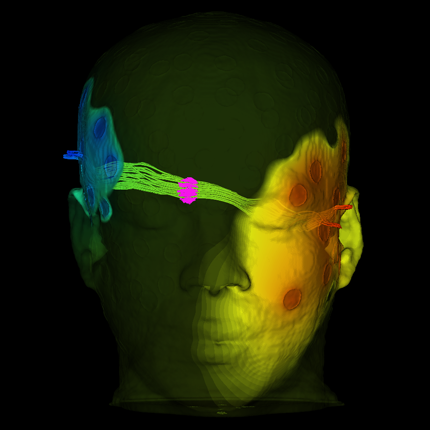
  A High-Resolution Head and Brain Computer Model for Forward and Inverse EEG Simulation A. Warner, J. Tate, B. Burton,, C.R. Johnson. In bioRxiv, Cold Spring Harbor Laboratory, Feb, 2019. DOI: 10.1101/552190 To conduct computational forward and inverse EEG studies of brain electrical activity, researchers must construct realistic head and brain computer models, which is both challenging and time consuming. The availability of realistic head models and corresponding imaging data is limited in terms of imaging modalities and patient diversity. In this paper, we describe a detailed head modeling pipeline and provide a high-resolution, multimodal, open-source, female head and brain model. The modeling pipeline specifically outlines image acquisition, preprocessing, registration, and segmentation; three-dimensional tetrahedral mesh generation; finite element EEG simulations; and visualization of the model and simulation results. The dataset includes both functional and structural images and EEG recordings from two high-resolution electrode configurations. The intermediate results and software components are also included in the dataset to facilitate modifications to the pipeline. This project will contribute to neuroscience research by providing a high-quality dataset that can be used for a variety of applications and a computational pipeline that may help researchers construct new head models more efficiently. |
  Skeletal Shape Correspondence through Entropy L. Tu, M. Styner, J. Vicory, S. Elhabian, R. Wang, J. Hong, B. Paniagua, J.C. Prieto, D. Yang, R. Whitaker, M. Pizer. In IEEE Transactions on Medical Imaging, Vol. 37, No. 1, IEEE, pp. 1--11. Jan, 2018. DOI: 10.1109/tmi.2017.2755550 We present a novel approach for improving the shape statistics of medical image objects by generating correspondence of skeletal points. Each object's interior is modeled by an s-rep, i.e., by a sampled, folded, two-sided skeletal sheet with spoke vectors proceeding from the skeletal sheet to the boundary. The skeleton is divided into three parts: the up side, the down side, and the fold curve. The spokes on each part are treated separately and, using spoke interpolation, are shifted along that skeleton in each training sample so as to tighten the probability distribution on those spokes' geometric properties while sampling the object interior regularly. As with the surface/boundary-based correspondence method of Cates et al., entropy is used to measure both the probability distribution tightness and the sampling regularity, here of the spokes' geometric properties. Evaluation on synthetic and real world lateral ventricle and hippocampus data sets demonstrate improvement in the performance of statistics using the resulting probability distributions. This improvement is greater than that achieved by an entropy-based correspondence method on the boundary points. |
  F. Mesadi, E. Erdil, M. Cetin, T. Tasdizen Image segmentation using disjunctive normal Bayesian shape, appearance models. In IEEE Transactions on Medical Imaging, Vol. 37, No. 1, IEEE, pp. 293--305. Jan, 2018. DOI: 10.1109/tmi.2017.2756929 The use of appearance and shape priors in image segmentation is known to improve accuracy; however, existing techniques have several drawbacks. For instance, most active shape and appearance models require landmark points and assume unimodal shape and appearance distributions, and the level set representation does not support construction of local priors. In this paper, we present novel appearance and shape models for image segmentation based on a differentiable implicit parametric shape representation called a disjunctive normal shape model (DNSM). The DNSM is formed by the disjunction of polytopes, which themselves are formed by the conjunctions of half-spaces. The DNSM's parametric nature allows the use of powerful local prior statistics, and its implicit nature removes the need to use landmarks and easily handles topological changes. In a Bayesian inference framework, we model arbitrary shape and appearance distributions using nonparametric density estimations, at any local scale. The proposed local shape prior results in accurate segmentation even when very few training shapes are available, because the method generates a rich set of shape variations by locally combining training samples. We demonstrate the performance of the framework by applying it to both 2-D and 3-D data sets with emphasis on biomedical image segmentation applications. |
  A virtual reality visualization tool for neuron tracing W Usher, P Klacansky, F Federer, PT Bremer, A Knoll, J. Yarch, A. Angelucci, V. Pascucci . In IEEE Transactions on Visualization and Computer Graphics, Vol. 24, No. 1, IEEE, pp. 994--1003. Jan, 2018. DOI: 10.1109/tvcg.2017.2744079 racing neurons in large-scale microscopy data is crucial to establishing a wiring diagram of the brain, which is needed to understand how neural circuits in the brain process information and generate behavior. Automatic techniques often fail for large and complex datasets, and connectomics researchers may spend weeks or months manually tracing neurons using 2D image stacks. We present a design study of a new virtual reality (VR) system, developed in collaboration with trained neuroanatomists, to trace neurons in microscope scans of the visual cortex of primates. We hypothesize that using consumer-grade VR technology to interact with neurons directly in 3D will help neuroscientists better resolve complex cases and enable them to trace neurons faster and with less physical and mental strain. We discuss both the design process and technical challenges in developing an interactive system to navigate and manipulate terabyte-sized image volumes in VR. Using a number of different datasets, we demonstrate that, compared to widely used commercial software, consumer-grade VR presents a promising alternative for scientists. |
  Neighbourhood looking glass: 360º automated characterisation of the built environment for neighbourhood effects research Q.C. Nguyen, M. Sajjadi, M. McCullough, M. Pham, T.T. Nguyen, W. Yu, H. Meng, M. Wen, F. Li, K.R. Smith, K. Brunisholz, T, Tasdizen. In Journal of Epidemiology and Community Health, BMJ, Jan, 2018. DOI: 10.1136/jech-2017-209456 Background |
  Domain adaptation for biomedical image segmentation using adversarial training M. Javanmardi, T. Tasdizen. In 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), IEEE, pp. 554-558. April, 2018. DOI: 10.1109/isbi.2018.8363637 Many biomedical image analysis applications require segmentation. Convolutional neural networks (CNN) have become a promising approach to segment biomedical images; however, the accuracy of these methods is highly dependent on the training data. We focus on biomedical image segmentation in the context where there is variation between source and target datasets and ground truth for the target dataset is very limited or non-existent. We use an adversarial based training approach to train CNNs to achieve good accuracy on the target domain. We use the DRIVE and STARE eye vasculture segmentation datasets and show that our approach can significantly improve results where we only use labels of one domain in training and test on the other domain. We also show improvements on membrane detection between MIC-CAI 2016 CREMI challenge and ISBI2013 EM segmentation challenge datasets. |
  Improving the robustness of convolutional networks to appearance variability in biomedical images T. Tasdizen, M. Sajjadi, M. Javanmardi, N. Ramesh. In 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), IEEE, April, 2018. DOI: 10.1109/isbi.2018.8363636 While convolutional neural networks (CNN) produce state-of-the-art results in many applications including biomedical image analysis, they are not robust to variability in the data that is not well represented by the training set. An important source of variability in biomedical images is the appearance of objects such as contrast and texture due to different imaging settings. We introduce the neighborhood similarity layer (NSL) which can be used in a CNN to improve robustness to changes in the appearance of objects that are not well represented by the training data. The proposed NSL transforms its input feature map at a given pixel by computing its similarity to the surrounding neighborhood. This transformation is spatially varying, hence not a convolution. It is differentiable; therefore, networks including the proposed layer can be trained in an end-to-end manner. We demonstrate the advantages of the NSL for the vasculature segmentation and cell detection problems. |
  Flexible Live‐Wire: Image Segmentation with Floating Anchors B. Summa, N. Faraj, C. Licorish, V. Pascucci. In Computer Graphics Forum, Vol. 37, No. 2, Wiley, pp. 321-328. May, 2018. DOI: 10.1111/cgf.13364 We introduce Flexible Live‐Wire, a generalization of the Live‐Wire interactive segmentation technique with floating anchors. In our approach, the user input for Live‐Wire is no longer limited to the setting of pixel‐level anchor nodes, but can use more general anchor sets. These sets can be of any dimension, size, or connectedness. The generality of the approach allows the design of a number of user interactions while providing the same functionality as the traditional Live‐Wire. In particular, we experiment with this new flexibility by designing four novel Live‐Wire interactions based on specific primitives: paint, pinch, probable, and pick anchors. These interactions are only a subset of the possibilities enabled by our generalization. Moreover, we discuss the computational aspects of this approach and provide practical solutions to alleviate any additional overhead. Finally, we illustrate our approach and new interactions through several example segmentations. |
  Nuclear proliferomics: A new field of study to identify signatures of nuclear materials as demonstrated on alpha-UO3 I. .J Schwerdt, A. Brenkmann, S. Martinson, B. D. Albrecht, S. Heffernan, M. R. Klosterman, T. Kirkham, T. Tasdizen, L. W. McDonald IV. In Talanta, Vol. 186, Elsevier BV, pp. 433--444. Aug, 2018. DOI: 10.1016/j.talanta.2018.04.092 The use of a limited set of signatures in nuclear forensics and nuclear safeguards may reduce the discriminating power for identifying unknown nuclear materials, or for verifying processing at existing facilities. Nuclear proliferomics is a proposed new field of study that advocates for the acquisition of large databases of nuclear material properties from a variety of analytical techniques. As demonstrated on a common uranium trioxide polymorph, α-UO3, in this paper, nuclear proliferomics increases the ability to improve confidence in identifying the processing history of nuclear materials. Specifically, α-UO3 was investigated from the calcination of unwashed uranyl peroxide at 350, 400, 450, 500, and 550 °C in air. Scanning electron microscopy (SEM) images were acquired of the surface morphology, and distinct qualitative differences are presented between unwashed and washed uranyl peroxide, as well as the calcination products from the unwashed uranyl peroxide at the investigated temperatures. Differential scanning calorimetry (DSC), UV–Vis spectrophotometry, powder X-ray diffraction (p-XRD), and thermogravimetric analysis-mass spectrometry (TGA-MS) were used to understand the source of these morphological differences as a function of calcination temperature. Additionally, the SEM images were manually segmented using Morphological Analysis for MAterials (MAMA) software to identify quantifiable differences in morphology for three different surface features present on the unwashed uranyl peroxide calcination products. No single quantifiable signature was sufficient to discern all calcination temperatures with a high degree of confidence; therefore, advanced statistical analysis was performed to allow the combination of a number of quantitative signatures, with their associated uncertainties, to allow for complete discernment by calcination history. Furthermore, machine learning was applied to the acquired SEM images to demonstrate automated discernment with at least 89% accuracy. |
  ISAVS: Interactive Scalable Analysis and Visualization System S. Petruzza, A. Venkat, A. Gyulassy, G. Scorzelli, F. Federer, A. Angelucci, V. Pascucci, P. T. Bremer. In ACM SIGGRAPH Asia 2017 Symposium on Visualization, ACM Press, 2017. DOI: 10.1145/3139295.3139299 Modern science is inundated with ever increasing data sizes as computational capabilities and image acquisition techniques continue to improve. For example, simulations are tackling ever larger domains with higher fidelity, and high-throughput microscopy techniques generate larger data that are fundamental to gather biologically and medically relevant insights. As the image sizes exceed memory, and even sometimes local disk space, each step in a scientific workflow is impacted. Current software solutions enable data exploration with limited interactivity for visualization and analytic tasks. Furthermore analysis on HPC systems often require complex hand-written parallel implementations of algorithms that suffer from poor portability and maintainability. We present a software infrastructure that simplifies end-to-end visualization and analysis of massive data. First, a hierarchical streaming data access layer enables interactive exploration of remote data, with fast data fetching to test analytics on subsets of the data. Second, a library simplifies the process of developing new analytics algorithms, allowing users to rapidly prototype new approaches and deploy them in an HPC setting. Third, a scalable runtime system automates mapping analysis algorithms to whatever computational hardware is available, reducing the complexity of developing scaling algorithms. We demonstrate the usability and performance of our system using a use case from neuroscience: filtering, registration, and visualization of tera-scale microscopy data. We evaluate the performance of our system using a leadership-class supercomputer, Shaheen II. |
 Longitudinal Modeling of Multi-modal Image Contrast Reveals Patterns of Early Brain Growth, A. Vardhan, J. Fishbaugh, C. Vachet, G. Gerig. In Medical Image Computing and Computer Assisted Intervention - MICCAI 2017, Springer International Publishing, pp. 75--83. 2017. DOI: 10.1007/978-3-319-66182-7_9 The brain undergoes rapid development during early childhood as a series of biophysical and chemical processes occur, which can be observed in magnetic resonance (MR) images as a change over time of white matter intensity relative to gray matter. Such a contrast change manifests in specific patterns in different imaging modalities, suggesting that brain maturation is encoded by appearance changes in multi-modal MRI. In this paper, we explore the patterns of early brain growth encoded by multi-modal contrast changes in a longitudinal study of children. For a given modality, contrast is measured by comparing histograms of intensity distributions between white and gray matter. Multivariate non-linear mixed effects (NLME) modeling provides subject-specific as well as population growth trajectories which accounts for contrast from multiple modalities. The multivariate NLME procedure and resulting non-linear contrast functions enable the study of maturation in various regions of interest. Our analysis of several brain regions in a study of 70 healthy children reveals a posterior to anterior pattern of timing of maturation in the major lobes of the cerebral cortex, with posterior regions maturing earlier than anterior regions. Furthermore, we find significant differences between maturation rates between males and females. |
  Neural circuitry at age 6~months associated with later repetitive behavior and sensory responsiveness in autism J. J. Wolff, M. R. Swanson, J. T. Elison, G. Gerig, J. R. Pruett, M. A. Styner, C. Vachet, K. N. Botteron, S. R. Dager, A. M. Estes, H. C. Hazlett, R. T. Schultz, M. D. Shen, L. Zwaigenbaum, J. Piven. In Molecular Autism, Vol. 8, No. 1, Springer Nature, March, 2017. DOI: 10.1186/s13229-017-0126-z Background |