Image Analysis
SCI's imaging work addresses fundamental questions in 2D and 3D image processing, including filtering, segmentation, surface reconstruction, and shape analysis. In low-level image processing, this effort has produce new nonparametric methods for modeling image statistics, which have resulted in better algorithms for denoising and reconstruction. Work with particle systems has led to new methods for visualizing and analyzing 3D surfaces. Our work in image processing also includes applications of advanced computing to 3D images, which has resulted in new parallel algorithms and real-time implementations on graphics processing units (GPUs). Application areas include medical image analysis, biological image processing, defense, environmental monitoring, and oil and gas.
Ross Whitaker
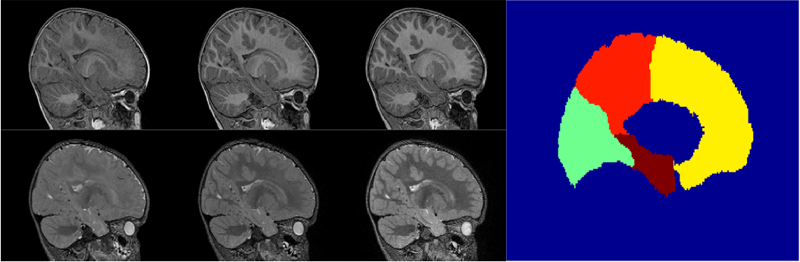
Segmentation
Chris Johnson
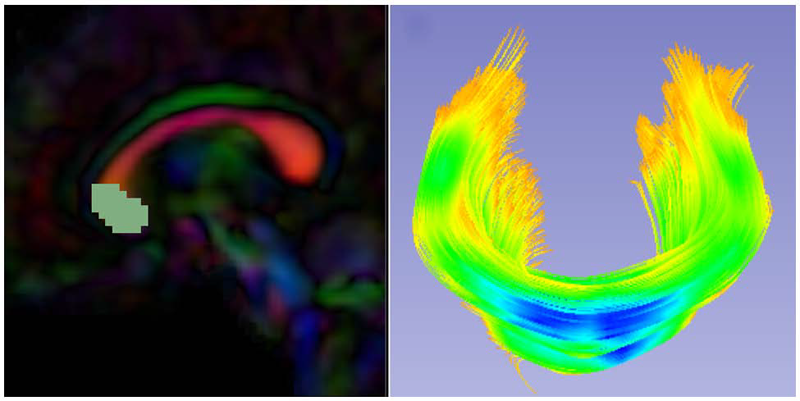
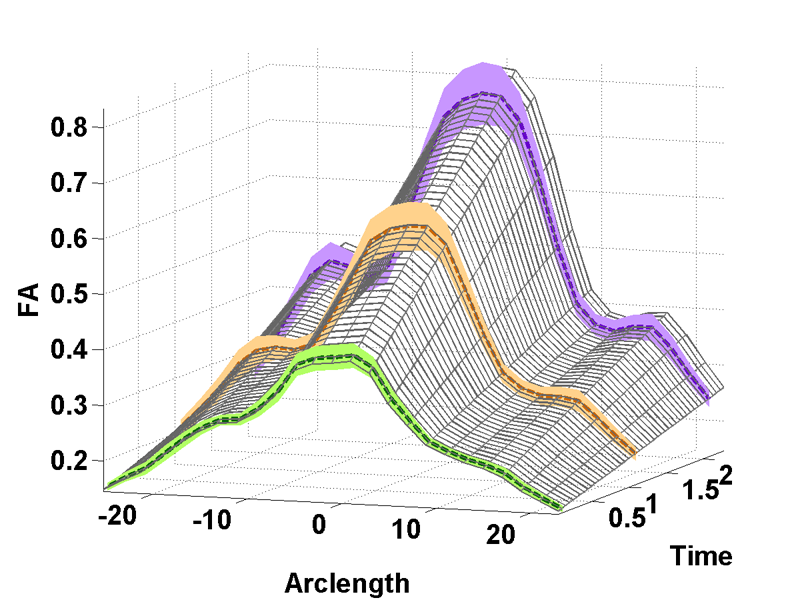
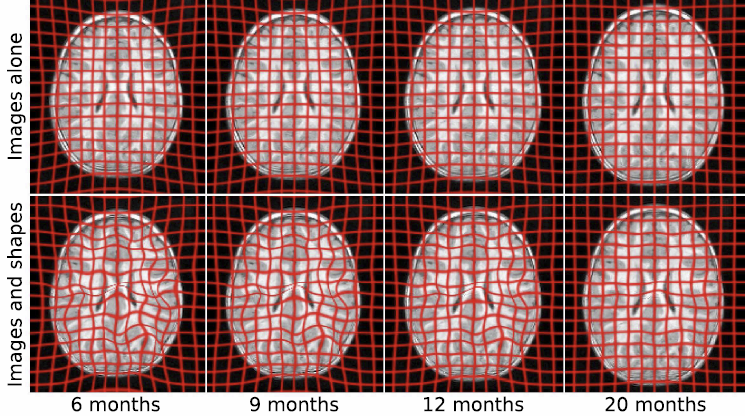
Diffusion Tensor AnalysisFunded Research Projects:
Publications in Image Analysis:
  Network inefficiencies in autism spectrum disorder at 24 months J.D. Lewis, A.C. Evans, J.R. Pruett, K. Botteron, L. Zwaigenbaum, A. Estes, G. Gerig, L. Collins, P. Kostopoulos, R. McKinstry, S. Dager, S. Paterson, R. Schultz, M. Styner, H. Hazlett, J. Piven, IBIS network. In Translational Psychiatry, Vol. 4, No. 5, Nature Publishing Group, pp. e388. May, 2014. DOI: 10.1038/tp.2014.24 Autism Spectrum Disorder (ASD) is a developmental disorder defined by behavioural symptoms that emerge during the first years of life. Associated with these symptoms are differences in the structure of a wide array of brain regions, and in the connectivity between these regions. However, the use of cohorts with large age variability and participants past the generally recognized age of onset of the defining behaviours means that many of the reported abnormalities may be a result of cascade effects of developmentally earlier deviations. This study assessed differences in connectivity in ASD at the age at which the defining behaviours first become clear. The participants were 113 24-month-olds at high risk for ASD, 31 of whom were classified as ASD, and 23 typically developing 24-month-olds at low risk for ASD. Utilizing diffusion data to obtain measures of the length and strength of connections between anatomical regions, we performed an analysis of network efficiency. Our results showed significantly decreased local and global efficiency over temporal, parietal, and occipital lobes in high-risk infants classified as ASD, relative to both low- and high-risk infants not classified as ASD. The frontal lobes showed only a reduction in global efficiency in Broca's area. Additionally, these same regions showed an inverse relation between efficiency and symptom severity across the high-risk infants. The results suggest delay or deficits in infants with ASD in the optimization of both local and global aspects of network structure in regions involved in processing auditory and visual stimuli, language, and nonlinguistic social stimuli. Keywords: autism, infant siblings, connectivity, network analysis, efficiency |
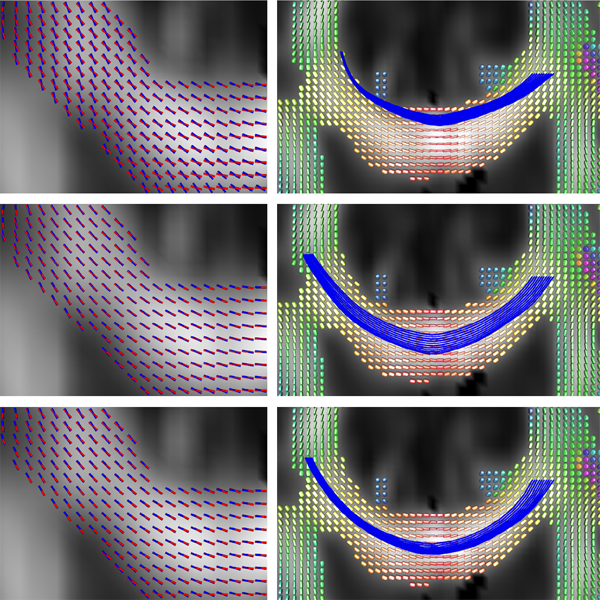
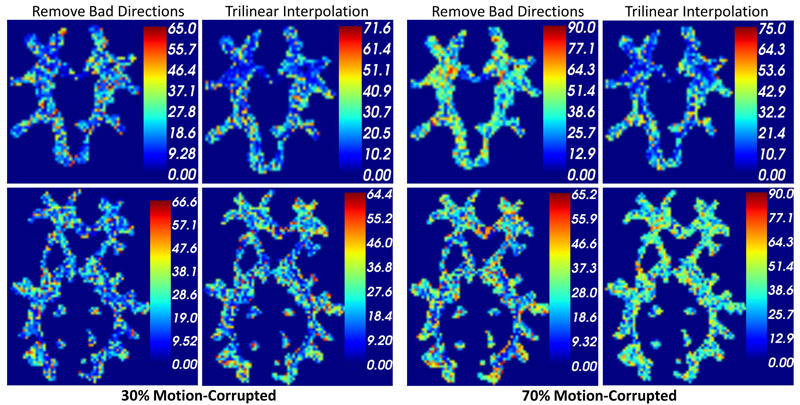
  Subject-Motion Correction in HARDI Acquisitions: Choices and Consequences, S. Elhabian, Y. Gur, J. Piven, M. Styner, I. Leppert, G.B. Pike, G. Gerig. In Proceeding of the 2014 Joint Annual Meeting ISMRM-ESMRMB, pp. (accepted). 2014. DOI: 10.3389/fneur.2014.00240 Unlike anatomical MRI where subject motion can most often be assessed by quick visual quality control, the detection, characterization and evaluation of the impact of motion in diffusion imaging are challenging issues due to the sensitivity of diffusion weighted imaging (DWI) to motion originating from vibration, cardiac pulsation, breathing and head movement. Post-acquisition motion correction is widely performed, e.g. using the open-source DTIprep software [1,2] or TORTOISE [3], but in particular in high angular resolution diffusion imaging (HARDI), users often do not fully understand the consequences of different types of correction schemes on the final analysis, and whether those choices may introduce confounding factors when comparing populations. Although there is excellent theoretical work on the number of directional DWI and its effect on the quality and crossing fiber resolution of orientation distribution functions (ODF), standard users lack clear guidelines and recommendations in practical settings. This research investigates motion correction using transformation and interpolation of affected DWI directions versus the exclusion of subsets of DWI’s, and its effects on diffusion measurements on the reconstructed fiber orientation diffusion functions and on the estimated fiber orientations. The various effects are systematically studied via a newly developed synthetic phantom and also on real HARDI data. |
  UNC-Utah NA-MIC framework for DTI fiber tract analysis A.R. Verde, F. Budin, J.-B. Berger, A. Gupta, M. Farzinfar, A. Kaiser, M. Ahn, H. Johnson, J. Matsui, H.C. Hazlett, A. Sharma, C. Goodlett, Y. Shi, S. Gouttard, C. Vachet, J. Piven, H. Zhu, G. Gerig, M. Styner. In Frontiers in Neuroinformatics, Vol. 7, No. 51, January, 2014. DOI: 10.3389/fninf.2013.00051 Diffusion tensor imaging has become an important modality in the field of neuroimaging to capture changes in micro-organization and to assess white matter integrity or development. While there exists a number of tractography toolsets, these usually lack tools for preprocessing or to analyze diffusion properties along the fiber tracts. Currently, the field is in critical need of a coherent end-to-end toolset for performing an along-fiber tract analysis, accessible to non-technical neuroimaging researchers. The UNC-Utah NA-MIC DTI framework represents a coherent, open source, end-to-end toolset for atlas fiber tract based DTI analysis encompassing DICOM data conversion, quality control, atlas building, fiber tractography, fiber parameterization, and statistical analysis of diffusion properties. Most steps utilize graphical user interfaces (GUI) to simplify interaction and provide an extensive DTI analysis framework for non-technical researchers/investigators. We illustrate the use of our framework on a small sample, cross sectional neuroimaging study of eight healthy 1-year-old children from the Infant Brain Imaging Study (IBIS) Network. In this limited test study, we illustrate the power of our method by quantifying the diffusion properties at 1 year of age on the genu and splenium fiber tracts. Keywords: neonatal neuroimaging, white matter pathways, magnetic resonance imaging, diffusion tensor imaging, diffusion imaging quality control, DTI atlas building |
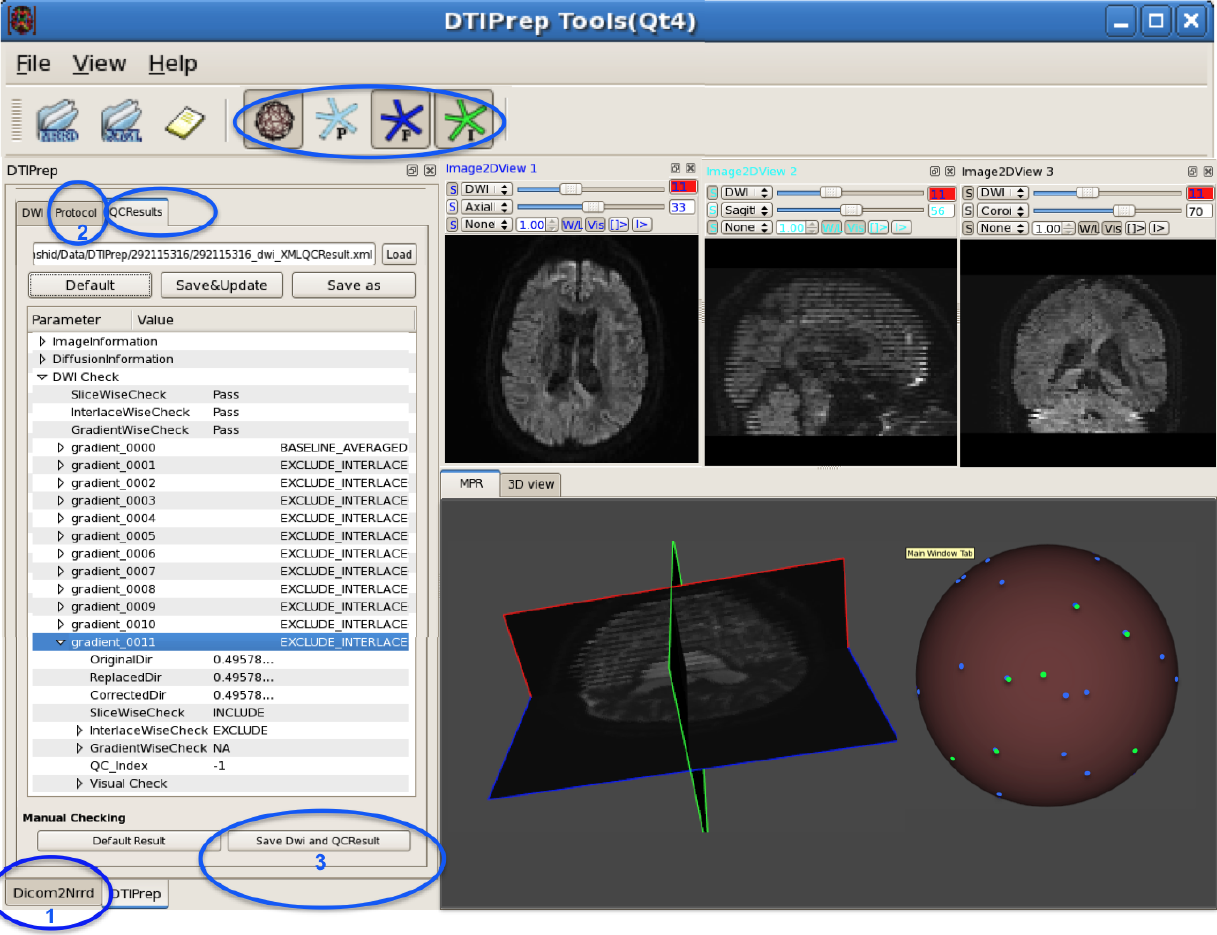
  DTIPrep: Quality Control of Diffusion-Weighted Images I. Oguz, M. Farzinfar, J. Matsui, F. Budin, Z. Liu, G. Gerig, H.J. Johnson, M.A. Styner. In Frontiers in Neuroinformatics, Vol. 8, No. 4, 2014. DOI: 10.3389/fninf.2014.00004 In the last decade, diffusion MRI (dMRI) studies of the human and animal brain have been used to investigate a multitude of pathologies and drug-related effects in neuroscience research. Study after study identifies white matter (WM) degeneration as a crucial biomarker for all these diseases. The tool of choice for studying WM is dMRI. However, dMRI has inherently low signal-to-noise ratio and its acquisition requires a relatively long scan time; in fact, the high loads required occasionally stress scanner hardware past the point of physical failure. As a result, many types of artifacts implicate the quality of diffusion imagery. Using these complex scans containing artifacts without quality control (QC) can result in considerable error and bias in the subsequent analysis, negatively affecting the results of research studies using them. However, dMRI QC remains an under-recognized issue in the dMRI community as there are no user-friendly tools commonly available to comprehensively address the issue of dMRI QC. As a result, current dMRI studies often perform a poor job at dMRI QC. Thorough QC of diffusion MRI will reduce measurement noise and improve reproducibility, and sensitivity in neuroimaging studies; this will allow researchers to more fully exploit the power of the dMRI technique and will ultimately advance neuroscience. Therefore, in this manuscript, we present our open-source software, DTIPrep, as a unified, user friendly platform for thorough quality control of dMRI data. These include artifacts caused by eddy-currents, head motion, bed vibration and pulsation, venetian blind artifacts, as well as slice-wise and gradient-wise intensity inconsistencies. This paper summarizes a basic set of features of DTIPrep described earlier and focuses on newly added capabilities related to directional artifacts and bias analysis. Keywords: diffusion MRI, Diffusion Tensor Imaging, Quality control, Software, open-source, preprocessing |
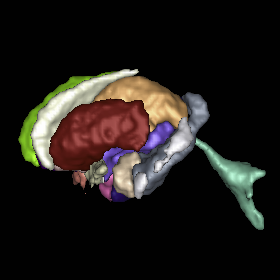
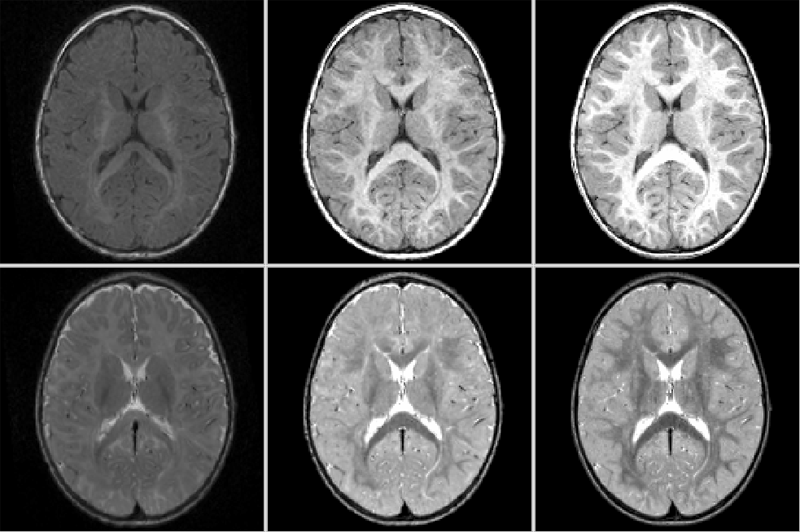
  Multi-atlas segmentation of subcortical brain structures via the AutoSeg software pipeline J. Wang, C. Vachet, A. Rumple, S. Gouttard, C. Ouzie, E. Perrot, G. Du, X. Huang, G. Gerig, M.A. Styner. In Frontiers in Neuroinformatics, Vol. 8, No. 7, 2014. DOI: 10.3389/fninf.2014.00007 Automated segmenting and labeling of individual brain anatomical regions, in MRI are challenging, due to the issue of individual structural variability. Although atlas-based segmentation has shown its potential for both tissue and structure segmentation, due to the inherent natural variability as well as disease-related changes in MR appearance, a single atlas image is often inappropriate to represent the full population of datasets processed in a given neuroimaging study. As an alternative for the case of single atlas segmentation, the use of multiple atlases alongside label fusion techniques has been introduced using a set of individual “atlases” that encompasses the expected variability in the studied population. In our study, we proposed a multi-atlas segmentation scheme with a novel graph-based atlas selection technique. We first paired and co-registered all atlases and the subject MR scans. A directed graph with edge weights based on intensity and shape similarity between all MR scans is then computed. The set of neighboring templates is selected via clustering of the graph. Finally, weighted majority voting is employed to create the final segmentation over the selected atlases. This multi-atlas segmentation scheme is used to extend a single-atlas-based segmentation toolkit entitled AutoSeg, which is an open-source, extensible C++ based software pipeline employing BatchMake for its pipeline scripting, developed at the Neuro Image Research and Analysis Laboratories of the University of North Carolina at Chapel Hill. AutoSeg performs N4 intensity inhomogeneity correction, rigid registration to a common template space, automated brain tissue classification based skull-stripping, and the multi-atlas segmentation. The multi-atlas-based AutoSeg has been evaluated on subcortical structure segmentation with a testing dataset of 20 adult brain MRI scans and 15 atlas MRI scans. The AutoSeg achieved mean Dice coefficients of 81.73% for the subcortical structures. Keywords: segmentation, Registration, MRI, Atlas, Brain, Insight Toolkit |
  Improved Segmentation of White Matter Tracts with Adaptive Riemannian Metrics X. Hao, K. Zygmunt, R.T. Whitaker, P.T. Fletcher. In Medical Image Analysis, Vol. 18, No. 1, pp. 161--175. Jan, 2014. DOI: 10.1016/j.media.2013.10.007 PubMed ID: 24211814 We present a novel geodesic approach to segmentation of white matter tracts from diffusion tensor imaging (DTI). Compared to deterministic and stochastic tractography, geodesic approaches treat the geometry of the brain white matter as a manifold, often using the inverse tensor field as a Riemannian metric. The white matter pathways are then inferred from the resulting geodesics, which have the desirable property that they tend to follow the main eigenvectors of the tensors, yet still have the flexibility to deviate from these directions when it results in lower costs. While this makes such methods more robust to noise, the choice of Riemannian metric in these methods is ad hoc. A serious drawback of current geodesic methods is that geodesics tend to deviate from the major eigenvectors in high-curvature areas in order to achieve the shortest path. In this paper we propose a method for learning an adaptive Riemannian metric from the DTI data, where the resulting geodesics more closely follow the principal eigenvector of the diffusion tensors even in high-curvature regions. We also develop a way to automatically segment the white matter tracts based on the computed geodesics. We show the robustness of our method on simulated data with different noise levels. We also compare our method with tractography methods and geodesic approaches using other Riemannian metrics and demonstrate that the proposed method results in improved geodesics and segmentations using both synthetic and real DTI data. Keywords: Conformal factor, Diffusion tensor imaging, Front-propagation, Geodesic, Riemannian manifold |
  Four‐dimensional tissue deformation reconstruction (4D TDR) validation using a real tissue phantom M. Szegedi, J. Hinkle, P. Rassiah, V. Sarkar, B. Wang, S. Joshi, B. Salter. In Journal of Applied Clinical Medical Physics, Vol. 14, No. 1, pp. 115-132. 2013. DOI: 10.1120/jacmp.v14i1.4012 Calculation of four‐dimensional (4D) dose distributions requires the remapping of dose calculated on each available binned phase of the 4D CT onto a reference phase for summation. Deformable image registration (DIR) is usually used for this task, but unfortunately almost always considers only endpoints rather than the whole motion path. A new algorithm, 4D tissue deformation reconstruction (4D TDR), that uses either CT projection data or all available 4D CT images to reconstruct 4D motion data, was developed. The purpose of this work is to verify the accuracy of the fit of this new algorithm using a realistic tissue phantom. A previously described fresh tissue phantom with implanted electromagnetic tracking (EMT) fiducials was used for this experiment. The phantom was animated using a sinusoidal and a real patient‐breathing signal. Four‐dimensional computer tomography (4D CT) and EMT tracking were performed. Deformation reconstruction was conducted using the 4D TDR and a modified 4D TDR which takes real tissue hysteresis (4D TDRHysteresis) into account. Deformation estimation results were compared to the EMT and 4D CT coordinate measurements. To eliminate the possibility of the high contrast markers driving the 4D TDR, a comparison was made using the original 4D CT data and data in which the fiducials were electronically masked. For the sinusoidal animation, the average deviation of the 4D TDR compared to the manually determined coordinates from 4D CT data was 1.9 mm, albeit with as large as 4.5 mm deviation. The 4D TDR calculation traces matched 95% of the EMT trace within 2.8 mm. The motion hysteresis generated by real tissue is not properly projected other than at endpoints of motion. Sinusoidal animation resulted in 95% of EMT measured locations to be within less than 1.2 mm of the measured 4D CT motion path, enabling accurate motion characterization of the tissue hysteresis. The 4D TDRHysteresis calculation traces accounted well for the hysteresis and matched 95% of the EMT trace within 1.6 mm. An irregular (in amplitude and frequency) recorded patient trace applied to the same tissue resulted in 95% of the EMT trace points within less than 4.5 mm when compared to both the 4D CT and 4D TDRHysteresis motion paths. The average deviation of 4D TDRHysteresis compared to 4D CT datasets was 0.9 mm under regular sinusoidal and 1.0 mm under irregular patient trace animation. The EMT trace data fit to the 4D TDRHysteresis was within 1.6 mm for sinusoidal and 4.5 mm for patient trace animation. While various algorithms have been validated for end‐to‐end accuracy, one can only be fully confident in the performance of a predictive algorithm if one looks at data along the full motion path. The 4D TDR, calculating the whole motion path rather than only phase‐ or endpoints, allows us to fully characterize the accuracy of a predictive algorithm, minimizing assumptions. This algorithm went one step further by allowing for the inclusion of tissue hysteresis effects, a real‐world effect that is neglected when endpoint‐only validation is performed. Our results show that the 4D TDRHysteresis correctly models the deformation at the endpoints and any intermediate points along the motion path. |
  Three-dimensional alignment and merging of confocal microscopy stacks N. Ramesh, T. Tasdizen. In 2013 IEEE International Conference on Image Processing, IEEE, September, 2013. DOI: 10.1109/icip.2013.6738297 We describe an efficient, robust, automated method for image alignment and merging of translated, rotated and flipped con-focal microscopy stacks. The samples are captured in both directions (top and bottom) to increase the SNR of the individual slices. We identify the overlapping region of the two stacks by using a variable depth Maximum Intensity Projection (MIP) in the z dimension. For each depth tested, the MIP images gives an estimate of the angle of rotation between the stacks and the shifts in the x and y directions using the Fourier Shift property in 2D. We use the estimated rotation angle, shifts in the x and y direction and align the images in the z direction. A linear blending technique based on a sigmoidal function is used to maximize the information from the stacks and combine them. We get maximum information gain as we combine stacks obtained from both directions. |
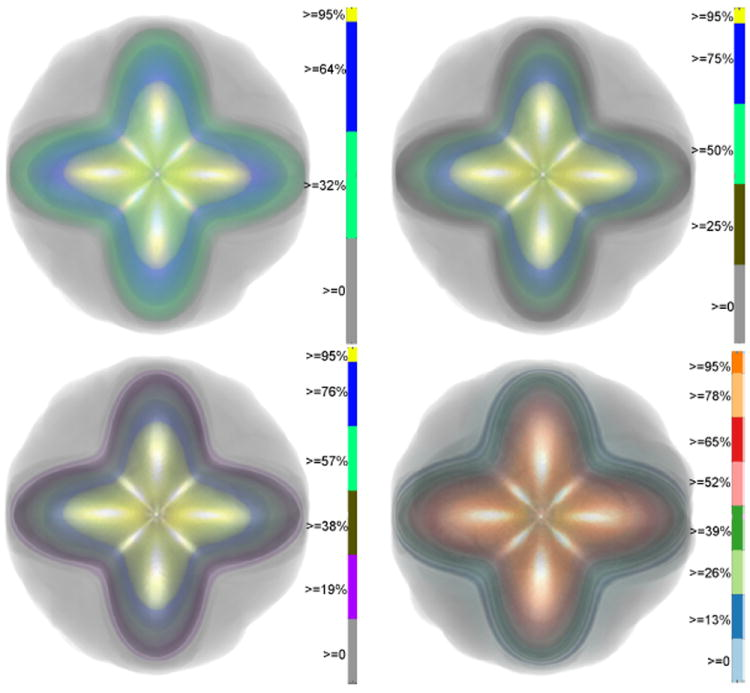
  Uncertainty Visualization in HARDI based on Ensembles of ODFs F. Jiao, J.M. Phillips, Y. Gur, C.R. Johnson. In Proceedings of 2013 IEEE Pacific Visualization Symposium, pp. 193--200. 2013. PubMed ID: 24466504 PubMed Central ID: PMC3898522 In this paper, we propose a new and accurate technique for uncertainty analysis and uncertainty visualization based on fiber orientation distribution function (ODF) glyphs, associated with high angular resolution diffusion imaging (HARDI). Our visualization applies volume rendering techniques to an ensemble of 3D ODF glyphs, which we call SIP functions of diffusion shapes, to capture their variability due to underlying uncertainty. This rendering elucidates the complex heteroscedastic structural variation in these shapes. Furthermore, we quantify the extent of this variation by measuring the fraction of the volume of these shapes, which is consistent across all noise levels, the certain volume ratio. Our uncertainty analysis and visualization framework is then applied to synthetic data, as well as to HARDI human-brain data, to study the impact of various image acquisition parameters and background noise levels on the diffusion shapes. |
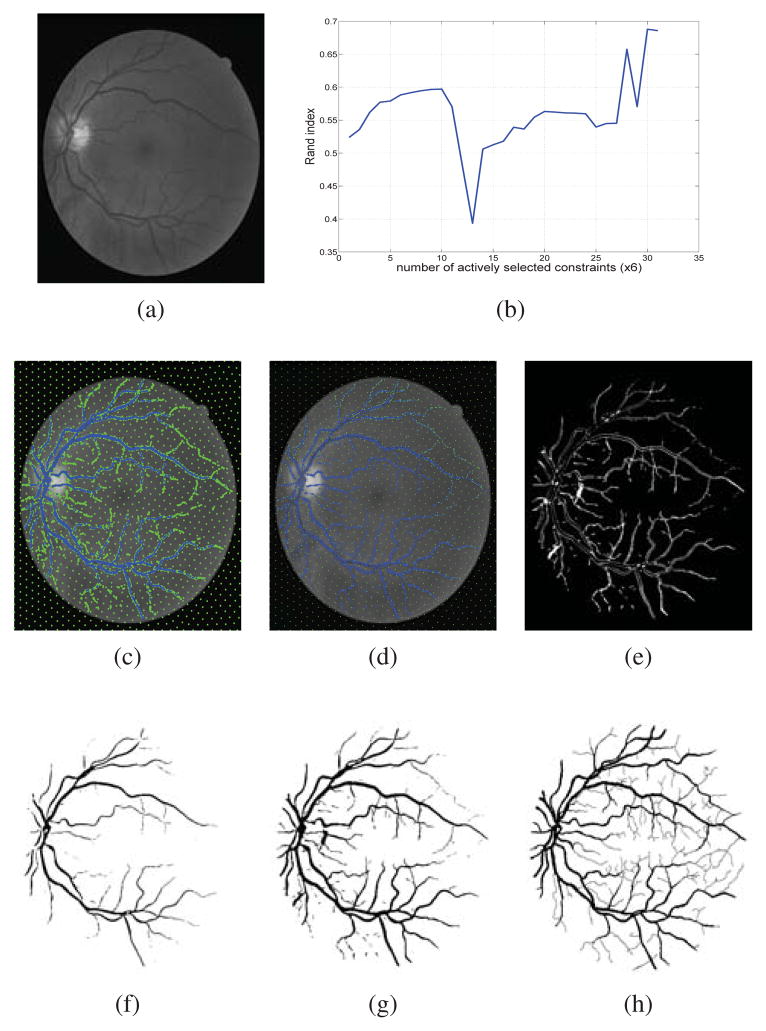
  Constrained Spectral Clustering for Image Segmentation J. Sourati, D.H. Brooks, J.G. Dy, E. Erdogmus. In IEEE International Workshop on Machine Learning for Signal Processing, pp. 1--6. 2013. DOI: 10.1109/MLSP Constrained spectral clustering with affinity propagation in its original form is not practical for large scale problems like image segmentation. In this paper we employ novelty selection sub-sampling strategy, besides using efficient numerical eigen-decomposition methods to make this algorithm work efficiently for images. In addition, entropy-based active learning is also employed to select the queries posed to the user more wisely in an interactive image segmentation framework. We evaluate the algorithm on general and medical images to show that the segmentation results will improve using constrained clustering even if one works with a subset of pixels. Furthermore, this happens more efficiently when pixels to be labeled are selected actively. |